Heterologous Expression of Serine Hydroxymethyltransferase-3 From Rice Confers Tolerance to Salinity Stress in E. coli and Arabidopsis
- PMID: 30941150
- PMCID: PMC6433796
- DOI: 10.3389/fpls.2019.00217
Heterologous Expression of Serine Hydroxymethyltransferase-3 From Rice Confers Tolerance to Salinity Stress in E. coli and Arabidopsis
Abstract
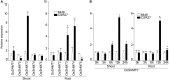
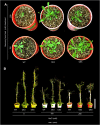
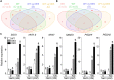
Among abiotic stresses, salt stress adversely affects growth and development in rice. Contrasting salt tolerant (CSR27), and salt sensitive (MI48) rice varieties provided information on an array of genes that may contribute for salt tolerance of rice. Earlier studies on transcriptome and proteome profiling led to the identification of salt stress-induced serine hydroxymethyltransferase-3 (SHMT3) gene. In the present study, the SHMT3 gene was isolated from salt-tolerant (CSR27) rice. OsSHMT3 exhibited salinity-stress induced accentuated and differential expression levels in different tissues of rice. OsSHMT3 was overexpressed in Escherichia coli and assayed for enzymatic activity and modeling protein structure. Further, Arabidopsis transgenic plants overexpressing OsSHMT3 exhibited tolerance toward salt stress. Comparative analyses of OsSHMT3 vis a vis wild type by ionomic, transcriptomic, and metabolic profiling, protein expression and analysis of various traits revealed a pivotal role of OsSHMT3 in conferring tolerance toward salt stress. The gene can further be used in developing gene-based markers for salt stress to be employed in marker assisted breeding programs.
Highlights: - The study provides information on mechanistic details of serine hydroxymethyl transferase gene for its salt tolerance in rice.
Keywords: Arabidopsis; aquaporins; glycine; rice; salinity; serine.
Figures



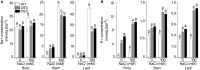
References
-
- Arzani A., Ashraf M. (2016). Smart engineering of genetic resources for enhanced salinity tolerance in crop plants. Crit. Rev. Plant Sci. 35 146–189. 10.1080/07352689 - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

