ABA-Induced Vegetative Diaspore Formation in Physcomitrella patens
- PMID: 30941155
- PMCID: PMC6433873
- DOI: 10.3389/fpls.2019.00315
ABA-Induced Vegetative Diaspore Formation in Physcomitrella patens
Abstract
The phytohormone abscisic acid (ABA) is a pivotal regulator of gene expression in response to various environmental stresses such as desiccation, salt and cold causing major changes in plant development and physiology. Here we show that in the moss Physcomitrella patens exogenous application of ABA triggers the formation of vegetative diaspores (brachycytes or brood cells) that enable plant survival in unfavorable environmental conditions. Such diaspores are round-shaped cells characterized by the loss of the central vacuole, due to an increased starch and lipid storage preparing these cells for growth upon suitable environmental conditions. To gain insights into the gene regulation underlying these developmental and physiological changes, we analyzed early transcriptome changes after 30, 60, and 180 min of ABA application and identified 1,030 differentially expressed genes. Among these, several groups can be linked to specific morphological and physiological changes during diaspore formation, such as genes involved in cell wall modifications. Furthermore, almost all members of ABA-dependent signaling and regulation were transcriptionally induced. Network analysis of transcription-associated genes revealed a large overlap of our study with ABA-dependent regulation in response to dehydration, cold stress, and UV-B light, indicating a fundamental function of ABA in diverse stress responses in moss. We also studied the evolutionary conservation of ABA-dependent regulation between moss and the seed plant Arabidopsis thaliana pointing to an early evolution of ABA-mediated stress adaptation during the conquest of the terrestrial habitat by plants.
Keywords: ABA; brachycyte; cell wall; diaspore; gene expression; moss.
Figures




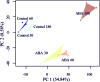

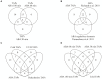
References
-
- Baldi P., Long A. D. (2001). A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17 509–519. - PubMed
LinkOut - more resources
Full Text Sources

