Gene flow signature in the S-allele region of cultivated buckwheat
- PMID: 30943914
- PMCID: PMC6448236
- DOI: 10.1186/s12870-019-1730-1
Gene flow signature in the S-allele region of cultivated buckwheat
Abstract
Background: Buckwheat (Fagopyrum esculentum Moench.) is an annual crop that originated in southern China. The nutritious seeds are used in cooking much like cereal grains. Buckwheat is an outcrossing species with heteromorphic self-incompatibility due to its dimorphic (i.e., short- and long-styled) flowers and intra-morph infertility. The floral morphology and intra-morph incompatibility are both determined by a single S locus. Plants with short-styled flowers are heterozygous (S/s) and plants with long-styled flowers are homozygous recessive (s/s) at this locus, and the S/S genotype is not found. Recently, we built a draft genome assembly of buckwheat and identified the 5.4-Mb-long S-allele region harbored by short-styled plants. In this study, the first report on the genome-wide diversity of buckwheat, we used a genotyping-by-sequencing (GBS) dataset to evaluate the genome-wide nucleotide diversity within cultivated buckwheat landraces worldwide. We also investigated the utility of the S-allele region for phylogenetic analysis of buckwheat.
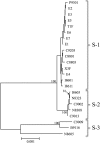
Results: Buckwheat showed high nucleotide diversity (0.0065), comparable to that of other outcrossing plants, based on a genome-wide simple nucleotide polymorphism (SNP) analysis. Phylogenetic analyses based on genome-wide SNPs showed that cultivated buckwheat comprises two groups, Asian and European, and revealed lower nucleotide diversity in the European group (0.0055) and low differentiation between the Asian and European groups. The nucleotide diversity (0.0039) estimated from SNPs in the S-allele region is lower than that in genome-wide SNPs. Phylogenetic analysis based on this region detected three diverged groups, S-1, S-2, and S-3.
Conclusion: The SNPs detected using the GBS dataset were effective for elucidating the evolutionary history of buckwheat, and led to the following conclusions: (1) the low nucleotide diversity of the entire genome in the European group and low differentiation between the Asian and European groups suggested genetic bottlenecks associated with dispersion from Asia to Europe, and/or recent intensified cultivation and selection in Europe; and (2) the high diversification in the S-allele region was indicative of gene flows from wild to cultivated buckwheat, suggesting that cultivated buckwheat may have multiple origins.
Keywords: Buckwheat; Crop evolution; GBS; Heteromorphic self-incompatibility.
Conflict of interest statement
Authors’ information
Laboratory of Crop Evolution, Division of Applied Bioscience, Graduate School of Agriculture, Kitashirakawa oiwake-cho, Sakyou-ku, Kyoto 606–8501, JAPAN.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


Similar articles
-
S-LOCUS EARLY FLOWERING 3 is exclusively present in the genomes of short-styled buckwheat plants that exhibit heteromorphic self-incompatibility.PLoS One. 2012;7(2):e31264. doi: 10.1371/journal.pone.0031264. Epub 2012 Feb 1. PLoS One. 2012. PMID: 22312442 Free PMC article.
-
Buckwheat heteromorphic self-incompatibility: genetics, genomics and application to breeding.Breed Sci. 2020 Mar;70(1):32-38. doi: 10.1270/jsbbs.19083. Epub 2020 Jan 30. Breed Sci. 2020. PMID: 32351302 Free PMC article.
-
Identification of a gene encoding polygalacturonase expressed specifically in short styles in distylous common buckwheat (Fagopyrum esculentum).Heredity (Edinb). 2019 Oct;123(4):492-502. doi: 10.1038/s41437-019-0227-x. Epub 2019 May 10. Heredity (Edinb). 2019. PMID: 31076649 Free PMC article.
-
Genetic and genomic research for the development of an efficient breeding system in heterostylous self-incompatible common buckwheat (Fagopyrum esculentum).Theor Appl Genet. 2020 May;133(5):1641-1653. doi: 10.1007/s00122-020-03572-6. Epub 2020 Mar 9. Theor Appl Genet. 2020. PMID: 32152716 Review.
-
Beyond the Cereal Box: Breeding Buckwheat as a Strategic Crop for Human Nutrition.Plant Foods Hum Nutr. 2021 Dec;76(4):399-409. doi: 10.1007/s11130-021-00930-7. Epub 2021 Oct 15. Plant Foods Hum Nutr. 2021. PMID: 34652552 Review.
Cited by
-
Comparison of Plant Morphology, Yield and Nutritional Quality of Fagopyrum esculentum and Fagopyrum tataricum Grown under Field Conditions in Belgium.Plants (Basel). 2021 Jan 28;10(2):258. doi: 10.3390/plants10020258. Plants (Basel). 2021. PMID: 33525666 Free PMC article.
-
History of the progressive development of genetic marker systems for common buckwheat.Breed Sci. 2020 Mar;70(1):13-18. doi: 10.1270/jsbbs.19075. Epub 2020 Jan 30. Breed Sci. 2020. PMID: 32351300 Free PMC article.
-
Breeding Buckwheat for Increased Levels of Rutin, Quercetin and Other Bioactive Compounds with Potential Antiviral Effects.Plants (Basel). 2020 Nov 24;9(12):1638. doi: 10.3390/plants9121638. Plants (Basel). 2020. PMID: 33255469 Free PMC article. Review.
-
Genomics-assisted breeding in minor and pseudo-cereals.Breed Sci. 2020 Mar;70(1):19-31. doi: 10.1270/jsbbs.19100. Epub 2020 Feb 1. Breed Sci. 2020. PMID: 32351301 Free PMC article.
-
FaesAP3_1 Regulates the FaesELF3 Gene Involved in Filament-Length Determination of Long-Homostyle Fagopyrum esculentum.Int J Mol Sci. 2022 Nov 19;23(22):14403. doi: 10.3390/ijms232214403. Int J Mol Sci. 2022. PMID: 36430880 Free PMC article.
References
-
- Ohnishi O. On the origin of cultivated common buckwheat based on allozyme analyses of cultivated and wild populations of common buckwheat. Fagopyrum. 2009;26:3–9.
-
- Bonafaccia G, Marocchini M, Kreft I. Composition and technological properties of the flour and bran from common and tartary buckwheat. Food Chem. 2003;80:9–15. doi: 10.1016/S0308-8146(02)00228-5. - DOI
-
- Ahmed A, Khalid N, Ahmed A, Abbasi NA, Latif MSZ, Randhawa MA. (2014) phytochemicals and biofunctional properties of buckwheat: a review. J Agric Sci. 2014;152:349–369. doi: 10.1017/S0021859613000166. - DOI
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

