Expression of C-terminal ALK, RET, or ROS1 in lung cancer cells with or without fusion
- PMID: 30943926
- PMCID: PMC6446279
- DOI: 10.1186/s12885-019-5527-2
Expression of C-terminal ALK, RET, or ROS1 in lung cancer cells with or without fusion
Abstract
Background: Genetic alterations, including mutation of epidermal growth factor receptor or v-Ki-ras2 kirsten rat sarcoma viral oncogene homolog and fusion of anaplastic lymphoma kinase (ALK), RET proto-oncogene (RET), or v-ros UR2 sarcoma virus oncogene homolog 1 (ROS1), occur in non-small cell lung cancers, and these oncogenic drivers are important biomarkers for targeted therapies. A useful technique to screen for these fusions is the detection of native carboxy-terminal (C-terminal) protein by immunohistochemistry; however, the effects of other genetic alterations on C-terminal expression is not fully understood. In this study, we evaluated whether C-terminal expression is specifically elevated by fusion with or without typical genetic alterations of lung cancer.
Methods: In 37 human lung cancer cell lines and four tissue specimens, protein and mRNA levels were measured by capillary western blotting and reverse transcription-PCR, respectively.
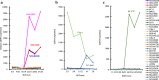
Results: Compared with the median of all 37 cell lines, mRNA levels at the C-terminus of all five of the fusion-positive cell lines tested (three ALK, one RET, and one ROS1) were elevated at least 2000-, 300-, or 2000-fold, respectively, and high C-terminal protein expression was detected. In an ALK fusion-positive tissue specimen, the mRNA and protein levels of C-terminal ALK were also markedly elevated. Meanwhile, in one of 36 RET fusion-negative cell lines, RET mRNA levels at the C-terminus were elevated at least 500-fold compared with the median of all 37 cell lines, and high C-terminal protein expression was detected despite the absence of RET fusion.
Conclusions: This study of 37 cell lines and four tissue specimens shows the detection of C-terminal ALK or ROS1 proteins could be a comprehensive method to determine ALK or ROS1 fusion, whereas not only the detection of C-terminal RET protein but also other methods would be needed to determine RET fusion.
Keywords: ALK; Alectinib; EML4; Fusion; Immunohistochemistry; Lung cancer; RET; ROS1; Rearrangement.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable because all materials were commercially available products. All studies were ethically reviewed and approved by the ethical review committee at Chugai Pharmaceutical Co., Ltd..
Consent for publication
Not applicable.
Competing interests
The authors are employees of Chugai Pharmaceutical Co., Ltd., and declare no conflict of interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
-
- Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, Fujiwara S, Watanabe H, Kurashina K, Hatanaka H, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448(7153):561–566. - PubMed
-
- Houtman SH, Rutteman M, De Zeeuw CI, French PJ. Echinoderm microtubule-associated protein like protein 4, a member of the echinoderm microtubule-associated protein family, stabilizes microtubules. Neuroscience. 2007;144(4):1373–1382. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

