hicap: In Silico Serotyping of the Haemophilus influenzae Capsule Locus
- PMID: 30944197
- PMCID: PMC6535587
- DOI: 10.1128/JCM.00190-19
hicap: In Silico Serotyping of the Haemophilus influenzae Capsule Locus
Abstract
Haemophilus influenzae exclusively colonizes the human nasopharynx and can cause a variety of respiratory infections as well as invasive diseases, including meningitis and sepsis. A key virulence determinant of H. influenzae is the polysaccharide capsule, of which six serotypes are known, each encoded by a distinct variation of the capsule biosynthesis locus (cap-a to cap-f). H. influenzae type b (Hib) was historically responsible for the majority of invasive H. influenzae disease, and its prevalence has been markedly reduced in countries that have implemented vaccination programs targeting this serotype. In the postvaccine era, nontypeable H. influenzae emerged as the most dominant group causing disease, but in recent years a resurgence of encapsulated H. influenzae strains has also been observed, most notably serotype a. Given the increasing incidence of encapsulated strains and the high frequency of Hib in countries without vaccination programs, there is growing interest in genomic epidemiology of H. influenzae Here we present hicap, a software tool for rapid in silico serotype prediction from H. influenzae genome sequences. hicap is written using Python3 and is freely available at https://github.com/scwatts/hicap under the GNU General Public License v3 (GPL3). To demonstrate the utility of hicap, we used it to investigate the cap locus diversity and distribution in 691 high-quality H. influenzae genomes from GenBank. These analyses identified cap loci in 95 genomes and confirmed the general association of each serotype with a unique clonal lineage, and they also identified occasional recombination between lineages that gave rise to hybrid cap loci (2% of encapsulated strains).
Keywords: Haemophilus influenzae; capsule; genomics; serotyping; surveillance.
Copyright © 2019 Watts and Holt.
Figures

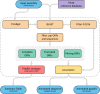
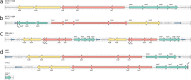

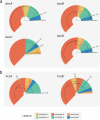

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

