Ca2+-dependent regulation of sodium channels NaV1.4 and NaV1.5 is controlled by the post-IQ motif
- PMID: 30944319
- PMCID: PMC6447637
- DOI: 10.1038/s41467-019-09570-7
Ca2+-dependent regulation of sodium channels NaV1.4 and NaV1.5 is controlled by the post-IQ motif
Abstract
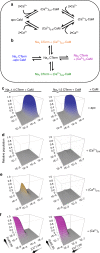
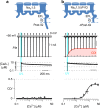
Skeletal muscle voltage-gated Na+ channel (NaV1.4) activity is subject to calmodulin (CaM) mediated Ca2+-dependent inactivation; no such inactivation is observed in the cardiac Na+ channel (NaV1.5). Taken together, the crystal structures of the NaV1.4 C-terminal domain relevant complexes and thermodynamic binding data presented here provide a rationale for this isoform difference. A Ca2+-dependent CaM N-lobe binding site previously identified in NaV1.5 is not present in NaV1.4 allowing the N-lobe to signal other regions of the NaV1.4 channel. Consistent with this mechanism, removing this binding site in NaV1.5 unveils robust Ca2+-dependent inactivation in the previously insensitive isoform. These findings suggest that Ca2+-dependent inactivation is effected by CaM's N-lobe binding outside the NaV C-terminal while CaM's C-lobe remains bound to the NaV C-terminal. As the N-lobe binding motif of NaV1.5 is a mutational hotspot for inherited arrhythmias, the contributions of mutation-induced changes in CDI to arrhythmia generation is an intriguing possibility.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

