L-Lactate dehydrogenase B may be a predictive marker for sensitivity to anti-EGFR monoclonal antibodies in colorectal cancer cell lines
- PMID: 30944657
- PMCID: PMC6444471
- DOI: 10.3892/ol.2019.10075
L-Lactate dehydrogenase B may be a predictive marker for sensitivity to anti-EGFR monoclonal antibodies in colorectal cancer cell lines
Abstract
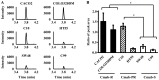
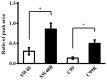
Recently, proteins derived from cancer cells have been widely investigated as biomarkers for predicting the efficacy of chemotherapy. In this study, to identify a sensitive biomarker for the efficacy of anti-epidermal growth factor receptor monoclonal antibodies (anti-EGFR mAbs), proteins derived from 6 colorectal cancer (CRC) cell lines with different sensitivities to cetuximab, an anti-EGFR mAb, were analyzed. Cytoplasmic and membrane proteins extracted from each CRC cell line were digested using trypsin and analyzed comprehensively using mass spectrometry. As a result, 148 and 146 peaks from cytoplasmic proteins and 363 and 267 peaks from membrane proteins were extracted as specific peaks for cetuximab-resistant and -sensitive CRC cell lines, respectively. By analyzing the proteins identified from the peptide peaks, cytoplasmic L-lactate dehydrogenase B (LDHB) was detected as a marker of cetuximab sensitivity, and it was confirmed that LDHB expression was increased in cetuximab-resistant CRC cell lines. Furthermore, LDHB expression levels were significantly upregulated with the acquisition of resistance to cetuximab in cetuximab-sensitive CRC cell lines. In conclusion, LDHB was identified as an important factor affecting cetuximab sensitivity using comprehensive proteome analysis for the first time.
Keywords: L-lactate dehydrogenase B; anti-epidermal growth factor receptor monoclonal antibodies; colorectal cancer; effect predictor; proteome.
Figures

References
-
- Gregorc V, Novello S, Lazzari C, Barni S, Aieta M, Mencoboni M, Grossi F, De Pas T, de Marinis F, Bearz A, et al. Predictive value of a proteomic signature in patients with non-small-cell lung cancer treated with second-line erlotinib or chemotherapy (PROSE): A biomarker-stratified, randomised phase 3 trial. Lancet Oncol. 2014;15:713–721. doi: 10.1016/S1470-2045(14)70162-7. - DOI - PubMed
-
- Lazzari C, Spreafico A, Bachi A, Roder H, Floriani I, Garavaglia D, Cattaneo A, Grigorieva J, Viganò MG, Sorlini C, et al. Changes in plasma mass-spectral profile in course of treatment of non-small cell lung cancer patients with epidermal growth factor receptor tyrosine kinase inhibitors. J Thorac Oncol. 2012;7:40–48. doi: 10.1097/JTO.0b013e3182307f17. - DOI - PubMed
-
- Garrisi VM, Bongarzone I, Mangia A, Cremona M, De Bortoli M, Vaghi E, Galetta D, Pastorino U, Quaranta M, Abbate I, Paradiso A. Characterization of a serum protein pattern from NSCLC patients treated with Gefitinib. Clin Biochem. 2011;44:936–940. doi: 10.1016/j.clinbiochem.2011.04.013. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
