Old wild wolves: ancient DNA survey unveils population dynamics in Late Pleistocene and Holocene Italian remains
- PMID: 30944772
- PMCID: PMC6441319
- DOI: 10.7717/peerj.6424
Old wild wolves: ancient DNA survey unveils population dynamics in Late Pleistocene and Holocene Italian remains
Abstract
Background: The contemporary Italian wolf (Canis lupus italicus) represents a case of morphological and genetic uniqueness. Today, Italian wolves are also the only documented population to fall exclusively within the mitochondrial haplogroup 2, which was the most diffused across Eurasian and North American wolves during the Late Pleistocene. However, the dynamics leading to such distinctiveness are still debated.
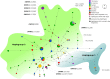
Methods: In order to shed light on the ancient genetic variability of this wolf population and on the origin of its current diversity, we collected 19 Late Pleistocene-Holocene samples from northern Italy, which we analyzed at a short portion of the hypervariable region 1 of the mitochondrial DNA, highly informative for wolf and dog phylogenetic analyses.
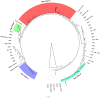
Results: Four out of the six detected haplotypes matched the ones found in ancient wolves from northern Europe and Beringia, or in modern European and Chinese wolves, and appeared closely related to the two haplotypes currently found in Italian wolves. The haplotype of two Late Pleistocene samples matched with primitive and contemporary dog sequences from the canine mitochondrial clade A. All these haplotypes belonged to haplogroup 2. The only exception was a Holocene sample dated 3,250 years ago, affiliated to haplogroup 1.
Discussion: In this study we describe the genetic variability of the most ancient wolf specimens from Italy analyzed so far, providing a preliminary overview of the genetic make-up of the population that inhabited this area from the last glacial maximum to the Middle Age period. Our results endorsed that the genetic diversity carried by the Pleistocene wolves here analyzed showed a strong continuity with other northern Eurasian wolf specimens from the same chronological period. Contrarily, the Holocene samples showed a greater similarity only with modern sequences from Europe and Asia, and the occurrence of an haplogroup 1 haplotype allowed to date back previous finding about its presence in this area. Moreover, the unexpected discovery of a 24,700-year-old sample carrying a haplotype that, from the fragment here obtained, falls within the canine clade A, could represent the oldest evidence in Europe of such dog-rich clade. All these findings suggest complex population dynamics that deserve to be further investigated based on mitochondrial or whole genome sequencing.
Keywords: Ancient DNA; Canid; Canis lupus; Control region; HVR1 variability; Italian wolf; Population genetics; Wolf; mtDNA.
Conflict of interest statement
Davide Palumbo is a zoologist consultant for Ente di Gestione per i Parchi e la Biodiversità Emilia Orientale. Marco Galaverni is an employee at the World Wildlife Fund (WWF), Conservation Unit—Italy.
Figures

References
-
- Aghbolaghi MA, Rezaei HR, Scandura M, Kaboli M. Low gene flow between Iranian Grey Wolves (Canis lupus) and dogs documented using uniparental genetic markers. Zoology in the Middle East. 2014;60(2):95–106. doi: 10.1080/09397140.2014.914708. - DOI
-
- Allentoft ME, Sikora M, Sjögren K-G, Rasmussen S, Rasmussen M, Stenderup J, Damgaard PB, Schroeder H, Ahlström T, Vinner L, Malaspinas A-S, Margaryan A, Higham T, Chivall D, Lynnerup N, Harvig L, Baron J, Casa PD, Dabrowski P, Duffy PR, Ebel AV, Epimakhov A, Frei K, Furmanek M, Gralak T, Gromov A, Gronkiewicz S, Grupe G, Hajdu T, Jarysz R, Khartanovich V, Khokhlov A, Kiss V, Kolář J, Kriiska A, Lasak I, Longhi C, McGlynn G, Merkevicius A, Merkyte I, Metspalu M, Mkrtchyan R, Moiseyev V, Paja L, Pálfi G, Pokutta D, Pospieszny L, Price TD, Saag L, Sablin M, Shishlina N, Smrčka V, Soenov VI, Szeverényi V, Tóth G, Trifanova SV, Varul L, Vicze M, Yepiskoposyan L, Zhitenev V, Orlando L, Sicheritz-Pontén T, Brunak S, Nielsen R, Kristiansen K, Willerslev E. Population genomics of Bronze Age Eurasia. Nature. 2015;522(7555):167–172. doi: 10.1038/nature14507. - DOI - PubMed
-
- Andersen LW, Harms V, Caniglia R, Czarnomska SD, Fabbri E, Jędrzejewska B, Kluth G, Madsen AB, Nowak C, Pertoldi C, Randi E, Reinhardt I, Stronen AV. Long-distance dispersal of a wolf, Canis lupus, in northwestern Europe. Mammal Research. 2015;60(2):163–168. doi: 10.1007/s13364-015-0220-6. - DOI
-
- Boggiano F, Ciofi C, Boitani L, Formia A, Grottoli L, Natali C, Ciucci P. Detection of an East European wolf haplotype puzzles mitochondrial DNA monomorphism of the Italian wolf population. Mammalian Biology. 2013;78(5):374–378. doi: 10.1016/j.mambio.2013.06.001. - DOI
-
- Botigué LR, Song S, Scheu A, Gopalan S, Pendleton AL, Oetjens M, Taravella AM, Seregély T, Zeeb-Lanz A, Arbogast RM, Bobo D, Daly K, Unterländer M, Burger J, Kidd JM, Veeramah KR. Ancient European dog genomes reveal continuity since the Early Neolithic. Nature Communications. 2017;8:16082. doi: 10.1038/ncomms16082. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

