PANGEA-HIV 2: Phylogenetics And Networks for Generalised Epidemics in Africa
- PMID: 30946141
- PMCID: PMC6629166
- DOI: 10.1097/COH.0000000000000542
PANGEA-HIV 2: Phylogenetics And Networks for Generalised Epidemics in Africa
Abstract
Purpose of review: The HIV epidemic in sub-Saharan Africa is far from being under control and the ambitious UNAIDS targets are unlikely to be met by 2020 as declines in per-capita incidence being largely offset by demographic trends. There is an increasing number of proven and specific HIV prevention tools, but little consensus on how best to deploy them.
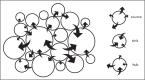
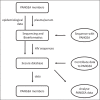
Recent findings: Traditionally, phylogenetics has been used in HIV research to reconstruct the history of the epidemic and date zoonotic infections, whereas more recent publications focus on HIV diversity and drug resistance. However, it is also the most powerful method of source attribution available for the study of HIV transmission. The PANGEA (Phylogenetics And Networks for Generalized Epidemics in Africa) consortium has generated over 18 000 NGS HIV sequences from five countries in sub-Saharan Africa. Using phylogenetic methods, we will identify characteristics of individuals or groups, which are most likely to be at risk of infection or at risk of infecting others.
Summary: Combining phylogenetics, phylodynamics and epidemiology will allow PANGEA to highlight where prevention efforts should be focussed to reduce the HIV epidemic most effectively. To maximise the public health benefit of the data, PANGEA offers accreditation to external researchers, allowing them to access the data and join the consortium. We also welcome submissions of other HIV sequences from sub-Saharan Africa to the database.
Figures

Similar articles
-
The Role of Phylogenetics in Discerning HIV-1 Mixing among Vulnerable Populations and Geographic Regions in Sub-Saharan Africa: A Systematic Review.Viruses. 2021 Jun 19;13(6):1174. doi: 10.3390/v13061174. Viruses. 2021. PMID: 34205246 Free PMC article.
-
Phylogenetic Tools for Generalized HIV-1 Epidemics: Findings from the PANGEA-HIV Methods Comparison.Mol Biol Evol. 2017 Jan;34(1):185-203. doi: 10.1093/molbev/msw217. Epub 2016 Oct 7. Mol Biol Evol. 2017. PMID: 28053012 Free PMC article.
-
HIV-1 full-genome phylogenetics of generalized epidemics in sub-Saharan Africa: impact of missing nucleotide characters in next-generation sequences.AIDS Res Hum Retroviruses. 2017 Nov;33(11):1083-1098. doi: 10.1089/AID.2017.0061. Epub 2017 May 25. AIDS Res Hum Retroviruses. 2017. PMID: 28540766 Free PMC article.
-
PANGEA-HIV: phylogenetics for generalised epidemics in Africa.Lancet Infect Dis. 2015 Mar;15(3):259-61. doi: 10.1016/S1473-3099(15)70036-8. Lancet Infect Dis. 2015. PMID: 25749217 Free PMC article. No abstract available.
-
Phylogenetic studies of transmission dynamics in generalized HIV epidemics: an essential tool where the burden is greatest?J Acquir Immune Defic Syndr. 2014 Oct 1;67(2):181-95. doi: 10.1097/QAI.0000000000000271. J Acquir Immune Defic Syndr. 2014. PMID: 24977473 Free PMC article. Review.
Cited by
-
Using mobile phone data to reveal risk flow networks underlying the HIV epidemic in Namibia.Nat Commun. 2021 May 14;12(1):2837. doi: 10.1038/s41467-021-23051-w. Nat Commun. 2021. PMID: 33990578 Free PMC article.
-
HIV-1 drug resistance genotyping success rates and correlates of Dried-blood spots and plasma specimen genotyping failure in a resource-limited setting.BMC Infect Dis. 2022 May 17;22(1):474. doi: 10.1186/s12879-022-07453-9. BMC Infect Dis. 2022. PMID: 35581555 Free PMC article.
-
The Role of Phylogenetics in Discerning HIV-1 Mixing among Vulnerable Populations and Geographic Regions in Sub-Saharan Africa: A Systematic Review.Viruses. 2021 Jun 19;13(6):1174. doi: 10.3390/v13061174. Viruses. 2021. PMID: 34205246 Free PMC article.
-
Evidence from HIV sequencing for blood-borne transmission in Africa.J Public Health Afr. 2025 Apr 30;16(1):715. doi: 10.4102/jphia.v16i1.715. eCollection 2025. J Public Health Afr. 2025. PMID: 40356728 Free PMC article. Review.
-
Human Immunodeficiency Virus (HIV) Genetic Diversity Informs Stage of HIV-1 Infection Among Patients Receiving Antiretroviral Therapy in Botswana.J Infect Dis. 2022 Apr 19;225(8):1330-1338. doi: 10.1093/infdis/jiab293. J Infect Dis. 2022. PMID: 34077517 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

