Genome-wide selection footprints and deleterious variations in young Asian allotetraploid rapeseed
- PMID: 30947395
- PMCID: PMC6737024
- DOI: 10.1111/pbi.13115
Genome-wide selection footprints and deleterious variations in young Asian allotetraploid rapeseed
Abstract
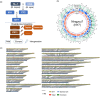
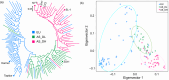
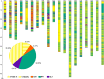
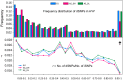
Brassica napus (AACC, 2n = 38) is an important oilseed crop grown worldwide. However, little is known about the population evolution of this species, the genomic difference between its major genetic groups, such as European and Asian rapeseed, and the impacts of historical large-scale introgression events on this young tetraploid. In this study, we reported the de novo assembly of the genome sequences of an Asian rapeseed (B. napus), Ningyou 7, and its four progenitors and compared these genomes with other available genomic data from diverse European and Asian cultivars. Our results showed that Asian rapeseed originally derived from European rapeseed but subsequently significantly diverged, with rapid genome differentiation after hybridization and intensive local selective breeding. The first historical introgression of B. rapa dramatically broadened the allelic pool but decreased the deleterious variations of Asian rapeseed. The second historical introgression of the double-low traits of European rapeseed (canola) has reshaped Asian rapeseed into two groups (double-low and double-high), accompanied by an increase in genetic load in the double-low group. This study demonstrates distinctive genomic footprints and deleterious SNP (single nucleotide polymorphism) variants for local adaptation by recent intra- and interspecies introgression events and provides novel insights for understanding the rapid genome evolution of a young allopolyploid crop.
Keywords: Asian rapeseed; allopolyploid; deleterious variations; introgression; selection footprints.
© 2019 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Figures


References
-
- Bancroft, I. , Morgan, C. , Fraser, F. , Higgins, J. , Wells, R. , Clissold, L. , Baker, D. et al. (2011) Dissecting the genome of the polyploid crop oilseed rape by transcriptome sequencing. Nat. Biotechnol. 29, 762–766. - PubMed
-
- Boetzer, M. , Henkel, C.V. , Jansen, H.J. , Butler, D. and Pirovano, W. (2011) Scaffolding pre‐assembled contigs using SSPACE. Bioinformatics, 27, 578–579. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

