Reversible induction of mitophagy by an optogenetic bimodular system
- PMID: 30948710
- PMCID: PMC6449392
- DOI: 10.1038/s41467-019-09487-1
Reversible induction of mitophagy by an optogenetic bimodular system
Abstract
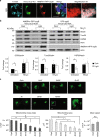
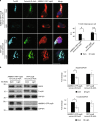
Autophagy-mediated degradation of mitochondria (mitophagy) is a key process in cellular quality control. Although mitophagy impairment is involved in several patho-physiological conditions, valuable methods to induce mitophagy with low toxicity in vivo are still lacking. Herein, we describe a new optogenetic tool to stimulate mitophagy, based on light-dependent recruitment of pro-autophagy protein AMBRA1 to mitochondrial surface. Upon illumination, AMBRA1-RFP-sspB is efficiently relocated from the cytosol to mitochondria, where it reversibly mediates mito-aggresome formation and reduction of mitochondrial mass. Finally, as a proof of concept of the biomedical relevance of this method, we induced mitophagy in an in vitro model of neurotoxicity, fully preventing cell death, as well as in human T lymphocytes and in zebrafish in vivo. Given the unique features of this tool, we think it may turn out to be very useful for a wide range of both therapeutic and research applications.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

