A NOTCH feed-forward loop drives reprogramming from adrenergic to mesenchymal state in neuroblastoma
- PMID: 30948783
- PMCID: PMC6449373
- DOI: 10.1038/s41467-019-09470-w
A NOTCH feed-forward loop drives reprogramming from adrenergic to mesenchymal state in neuroblastoma
Abstract
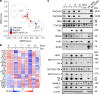
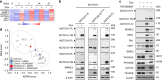
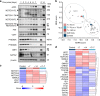
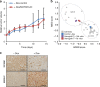
Transition between differentiation states in development occurs swift but the mechanisms leading to epigenetic and transcriptional reprogramming are poorly understood. The pediatric cancer neuroblastoma includes adrenergic (ADRN) and mesenchymal (MES) tumor cell types, which differ in phenotype, super-enhancers (SEs) and core regulatory circuitries. These cell types can spontaneously interconvert, but the mechanism remains largely unknown. Here, we unravel how a NOTCH3 intracellular domain reprogrammed the ADRN transcriptional landscape towards a MES state. A transcriptional feed-forward circuitry of NOTCH-family transcription factors amplifies the NOTCH signaling levels, explaining the swift transition between two semi-stable cellular states. This transition induces genome-wide remodeling of the H3K27ac landscape and a switch from ADRN SEs to MES SEs. Once established, the NOTCH feed-forward loop maintains the induced MES state. In vivo reprogramming of ADRN cells shows that MES and ADRN cells are equally oncogenic. Our results elucidate a swift transdifferentiation between two semi-stable epigenetic cellular states.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

