Evaluation of CD33 as a genetic risk factor for Alzheimer's disease
- PMID: 30949760
- PMCID: PMC7035471
- DOI: 10.1007/s00401-019-02000-4
Evaluation of CD33 as a genetic risk factor for Alzheimer's disease
Abstract
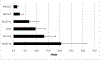
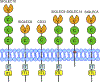
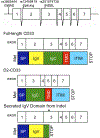
In 2011, genome-wide association studies implicated a polymorphism near CD33 as a genetic risk factor for Alzheimer's disease. This finding sparked interest in this member of the sialic acid-binding immunoglobulin-type lectin family which is linked to innate immunity. Subsequent studies found that CD33 is expressed in microglia in the brain and then investigated the molecular mechanism underlying the CD33 genetic association with Alzheimer's disease. The allele that protects from Alzheimer's disease acts predominately to increase a CD33 isoform lacking exon 2 at the expense of the prototypic, full-length CD33 that contains exon 2. Since this exon encodes the sialic acid ligand-binding domain, the finding that the loss of exon 2 was associated with decreased Alzheimer's disease risk was interpreted as meaning that a decrease in functional CD33 and its associated immune suppression was protective from Alzheimer's disease. However, this interpretation may need to be reconsidered given current findings that a genetic deletion which abrogates CD33 is not associated with Alzheimer's disease risk. Therefore, integrating currently available findings leads us to propose a model wherein the CD33 isoform lacking the ligand-binding domain represents a gain of function variant that reduces Alzheimer's disease risk.
Keywords: Alzheimer’s disease; CD33; Immunoreceptor tyrosine-based activation motif; Immunoreceptor tyrosine-based inhibitory motif; Inflammation; Molecular genetics; RNA splicing; SIGLEC.
Figures



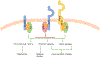
Comment in
-
Controversies about the subcellular localization and mechanisms of action of the Alzheimer's disease-protective CD33 splice variant.Acta Neuropathol. 2019 Oct;138(4):671-672. doi: 10.1007/s00401-019-02065-1. Epub 2019 Aug 22. Acta Neuropathol. 2019. PMID: 31440824 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

