Emerging views of mitophagy in immunity and autoimmune diseases
- PMID: 30951392
- PMCID: PMC6984455
- DOI: 10.1080/15548627.2019.1603547
Emerging views of mitophagy in immunity and autoimmune diseases
Abstract
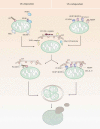
Mitophagy is a vital form of autophagy for selective removal of dysfunctional or redundant mitochondria. Accumulating evidence implicates elimination of dysfunctional mitochondria as a powerful means employed by autophagy to keep the immune system in check. The process of mitophagy may restrict inflammatory cytokine secretion and directly regulate mitochondrial antigen presentation and immune cell homeostasis. In this review, we describe distinctive pathways of mammalian mitophagy and highlight recent advances relevant to its function in immunity. In addition, we further discuss the direct and indirect evidence linking mitophagy to inflammation and autoimmunity underlying the pathogenesis of autoimmune diseases including inflammatory bowel diseases (IBD), systemic lupus erythematosus (SLE) and primary biliary cirrhosis (PBC).Abbreviations: AICD: activation induced cell death; AIM2: absent in melanoma 2; ALPL/HOPS: alkaline phosphatase, biomineralization associated; AMA: anti-mitochondrial antibodies; AMFR: autocrine motility factor receptor; ATG: autophagy-related; BCL2L13: BCL2 like 13; BNIP3: BCL2 interacting protein 3; BNIP3L/NIX: BCL2 interacting protein 3 like; CALCOCO2/NDP52: calcium binding and coiled-coil domain 2; CARD: caspase recruitment domain containing; CASP1: caspase 1; CD: Crohn disease; CGAS: cyclic GMP-AMP synthase; CXCL1: C-X-C motif chemokine ligand 1; DEN: diethylnitrosamine; DLAT/PDC-E2: dihydrolipoamide S-acetyltransferase; DNM1L/Drp1: dynamin 1 like; ESCRT: endosomal sorting complexes required for transport; FKBP8: FKBP prolyl isomerase 8; FUNDC1: Fun14 domain containing 1; GABARAP: GABA type A receptor-associated protein; HMGB1: high mobility group box 1; HPIV3: human parainfluenza virus type 3; IBD: inflammatory bowel diseases; IEC: intestinal epithelial cell; IFN: interferon; IL1B/IL-1β: interleukin 1 beta; iNK: invariant natural killer; IRGM: immunity related GTPase M; LIR: LC3-interacting region; LPS: lipopolysaccharide; LRRK2: leucine rich repeat kinase 2; MAP1LC3/LC3: microtubule associated protein 1 light chain 3; MARCH5: membrane associated ring-CH-type finger 5; MAVS: mitochondrial antiviral signaling protein; MDV: mitochondria-derived vesicle; MFN1: mitofusin 1; MHC: major histocompatibility complex; MIF: macrophage migration inhibitory factor; mtAP: mitochondrial antigen presentation; mtDNA: mitochondrial DNA; MTOR: mechanistic target of rapamycin kinase; mtROS: mitochondrial ROS; MUL1: mitochondrial E3 ubiquitin protein ligase 1; NBR1: NBR1 autophagy cargo receptor; NFKB/NF-ĸB: nuclear factor kappa B subunit; NK: natural killer; NLR: NOD-like receptor; NLRC4: NLR family CARD domain containing 4; NLRP3: NLR family pyrin domain containing 3; OGDH: oxoglutarate dehydrogenase; OMM: outer mitochondrial membrane; OPTN: optineurin; ox: oxidized; PARK7: Parkinsonism associated deglycase; PBC: primary biliary cirrhosis; PEX13: peroxisomal biogenesis factor 13; PHB/PHB1: prohibitin; PHB2: prohibitin 2; PIK3C3/VPS34: phosphatidylinositol 3-kinase catalytic subunit type 3; PINK1: PTEN induced kinase 1; PLEKHM1: pleckstrin homology and RUN domain containing M1; PRKN/PARK2: parkin RBR E3 ubiquitin protein ligase; RAB: member RAS oncogene family; RHEB: Ras homolog: mTORC1 binding; RIPK2: receptor interacting serine/threonine kinase 2; RLR: DDX58/RIG-I like receptor; ROS: reactive oxygen species; SBD: small bile ducts; SLC2A1/GLUT1: solute carrier family 2 member 1; SLE: systemic lupus erythematosus; SMURF1: SMAD specific E3 ubiquitin protein ligase 1; SQSTM1/p62: sequestosome 1; TAX1BP1: Tax1 binding protein 1; TCR: T cell receptor; TFAM: transcription factor A: mitochondrial; Th17: T helper 17; TLR9: toll like receptor 9; TMEM173/STING: transmembrane protein 173; TNF/TNF-α: tumor necrosis factor; Ub: ubiquitin; UC: ulcerative colitis; ULK1: unc-51 like autophagy activating kinase 1; WIPI: WD repeat domain: phosphoinositide interacting; ZFYVE1/DFCP1: zinc finger FYVE-type containing 1.
Keywords: Immune cells; inflammatory bowel diseases; inflammatory cytokines; mitophagy; primary biliary cirrhosis; systemic lupus erythematosus.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
