Small RNA Sequencing across Diverse Biofluids Identifies Optimal Methods for exRNA Isolation
- PMID: 30951671
- PMCID: PMC6557167
- DOI: 10.1016/j.cell.2019.03.024
Small RNA Sequencing across Diverse Biofluids Identifies Optimal Methods for exRNA Isolation
Abstract
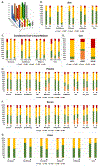
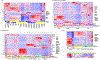
Poor reproducibility within and across studies arising from lack of knowledge regarding the performance of extracellular RNA (exRNA) isolation methods has hindered progress in the exRNA field. A systematic comparison of 10 exRNA isolation methods across 5 biofluids revealed marked differences in the complexity and reproducibility of the resulting small RNA-seq profiles. The relative efficiency with which each method accessed different exRNA carrier subclasses was determined by estimating the proportions of extracellular vesicle (EV)-, ribonucleoprotein (RNP)-, and high-density lipoprotein (HDL)-specific miRNA signatures in each profile. An interactive web-based application (miRDaR) was developed to help investigators select the optimal exRNA isolation method for their studies. miRDar provides comparative statistics for all expressed miRNAs or a selected subset of miRNAs in the desired biofluid for each exRNA isolation method and returns a ranked list of exRNA isolation methods prioritized by complexity, expression level, and reproducibility. These results will improve reproducibility and stimulate further progress in exRNA biomarker development.
Keywords: extracellular RNA; extracellular vesicles; lipoprotein; ribonucleoprotein.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures





Comment in
-
Mapping Extracellular RNA Sheds Lights on Distinct Carriers.Cell. 2019 Apr 4;177(2):228-230. doi: 10.1016/j.cell.2019.03.027. Cell. 2019. PMID: 30951666
References
-
- Beltrami C, Besnier M, Shantikumar S, Shearn AI, Rajakaruna C, Laftah A, Sessa F, Spinetti G, Petretto E, Angelini GD, et al. (2017). Human Pericardial Fluid Contains Exosomes Enriched with Cardiovascular-Expressed MicroRNAs and Promotes Therapeutic Angiogenesis. Mol Ther 25, 679–693. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL122547/HL/NHLBI NIH HHS/United States
- P50 CA217685/CA/NCI NIH HHS/United States
- UH3 TR000943/TR/NCATS NIH HHS/United States
- U54 DA036134/DA/NIDA NIH HHS/United States
- R01 HL133575/HL/NHLBI NIH HHS/United States
- R01 CA217833/CA/NCI NIH HHS/United States
- U19 CA179563/CA/NCI NIH HHS/United States
- UH3 TR000901/TR/NCATS NIH HHS/United States
- UH3 TR000890/TR/NCATS NIH HHS/United States
- U19 CA179512/CA/NCI NIH HHS/United States
- UH2 TR000901/TR/NCATS NIH HHS/United States
- K23 HL127099/HL/NHLBI NIH HHS/United States
- UH3 TR000906/TR/NCATS NIH HHS/United States
- P01 CA069246/CA/NCI NIH HHS/United States
- R35 CA232103/CA/NCI NIH HHS/United States
- UH2 TR000884/TR/NCATS NIH HHS/United States
- UH2 TR000906/TR/NCATS NIH HHS/United States
- T32 HD007203/HD/NICHD NIH HHS/United States
- UH2 TR000943/TR/NCATS NIH HHS/United States
- UH2 TR000890/TR/NCATS NIH HHS/United States
- R35 CA209904/CA/NCI NIH HHS/United States
- U01 HL126494/HL/NHLBI NIH HHS/United States
- UH3 TR000884/TR/NCATS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

