FoxK1 and FoxK2 in insulin regulation of cellular and mitochondrial metabolism
- PMID: 30952843
- PMCID: PMC6450906
- DOI: 10.1038/s41467-019-09418-0
FoxK1 and FoxK2 in insulin regulation of cellular and mitochondrial metabolism
Abstract
A major target of insulin signaling is the FoxO family of Forkhead transcription factors, which translocate from the nucleus to the cytoplasm following insulin-stimulated phosphorylation. Here we show that the Forkhead transcription factors FoxK1 and FoxK2 are also downstream targets of insulin action, but that following insulin stimulation, they translocate from the cytoplasm to nucleus, reciprocal to the translocation of FoxO1. FoxK1/FoxK2 translocation to the nucleus is dependent on the Akt-mTOR pathway, while its localization to the cytoplasm in the basal state is dependent on GSK3. Knockdown of FoxK1 and FoxK2 in liver cells results in upregulation of genes related to apoptosis and down-regulation of genes involved in cell cycle and lipid metabolism. This is associated with decreased cell proliferation and altered mitochondrial fatty acid metabolism. Thus, FoxK1/K2 are reciprocally regulated to FoxO1 following insulin stimulation and play a critical role in the control of apoptosis, metabolism and mitochondrial function.
Conflict of interest statement
The authors declare no competing interests.
Figures

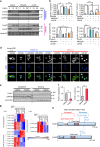



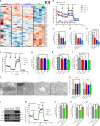

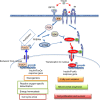
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

