N-terminal syndecan-2 domain selectively enhances 6-O heparan sulfate chains sulfation and promotes VEGFA165-dependent neovascularization
- PMID: 30952866
- PMCID: PMC6450910
- DOI: 10.1038/s41467-019-09605-z
N-terminal syndecan-2 domain selectively enhances 6-O heparan sulfate chains sulfation and promotes VEGFA165-dependent neovascularization
Erratum in
-
Publisher Correction: N-terminal syndecan-2 domain selectively enhances 6-O heparan sulfate chains sulfation and promotes VEGFA165-dependent neovascularization.Nat Commun. 2019 May 7;10(1):2124. doi: 10.1038/s41467-019-10205-0. Nat Commun. 2019. PMID: 31064993 Free PMC article.
Abstract
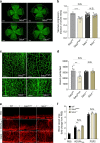
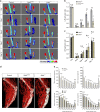
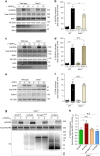
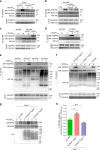
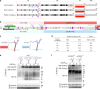
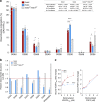
The proteoglycan Syndecan-2 (Sdc2) has been implicated in regulation of cytoskeleton organization, integrin signaling and developmental angiogenesis in zebrafish. Here we report that mice with global and inducible endothelial-specific deletion of Sdc2 display marked angiogenic and arteriogenic defects and impaired VEGFA165 signaling. No such abnormalities are observed in mice with deletion of the closely related Syndecan-4 (Sdc4) gene. These differences are due to a significantly higher 6-O sulfation level in Sdc2 versus Sdc4 heparan sulfate (HS) chains, leading to an increase in VEGFA165 binding sites and formation of a ternary Sdc2-VEGFA165-VEGFR2 complex which enhances VEGFR2 activation. The increased Sdc2 HS chains 6-O sulfation is driven by a specific N-terminal domain sequence; the insertion of this sequence in Sdc4 N-terminal domain increases 6-O sulfation of its HS chains and promotes Sdc2-VEGFA165-VEGFR2 complex formation. This demonstrates the existence of core protein-determined HS sulfation patterns that regulate specific biological activities.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

