Pellino1 regulates reversible ATM activation via NBS1 ubiquitination at DNA double-strand breaks
- PMID: 30952868
- PMCID: PMC6450972
- DOI: 10.1038/s41467-019-09641-9
Pellino1 regulates reversible ATM activation via NBS1 ubiquitination at DNA double-strand breaks
Abstract
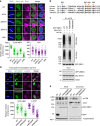
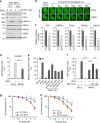
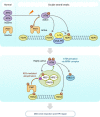
DNA double-strand break (DSB) signaling and repair are critical for genome integrity. They rely on highly coordinated processes including posttranslational modifications of proteins. Here we show that Pellino1 (Peli1) is a DSB-responsive ubiquitin ligase required for the accumulation of DNA damage response proteins and efficient homologous recombination (HR) repair. Peli1 is activated by ATM-mediated phosphorylation. It is recruited to DSB sites in ATM- and γH2AX-dependent manners. Interaction of Peli1 with phosphorylated histone H2AX enables it to bind to and mediate the formation of K63-linked ubiquitination of NBS1, which subsequently results in feedback activation of ATM and promotes HR repair. Collectively, these results provide a DSB-responsive factor underlying the connection between ATM kinase and DSB-induced ubiquitination.
Conflict of interest statement
The authors declare no competing interests.
Figures







References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

