O-GlcNAc Transferase Inhibition Differentially Affects Breast Cancer Subtypes
- PMID: 30952976
- PMCID: PMC6450885
- DOI: 10.1038/s41598-019-42153-6
O-GlcNAc Transferase Inhibition Differentially Affects Breast Cancer Subtypes
Abstract
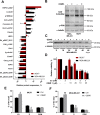
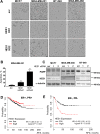
Post-translational modification of intracellular proteins with a single N-acetylglucosamine sugar (O-GlcNAcylation) regulates signaling, proliferation, metabolism and protein stability. In breast cancer, expression of the enzyme that catalyzes O-GlcNAcylation - O-GlcNAc-transferase (OGT), and the extent of protein O-GlcNAcylation, are upregulated in tumor tissue, and correlate with cancer progression. Here we compare the significance of O-GlcNAcylation in a panel of breast cancer cells of different phenotypes. We find a greater dependency on OGT among triple-negative breast cancer (TNBC) cell lines, which respond to OGT inhibition by undergoing cell cycle arrest and apoptosis. Searching for the cause of this response, we evaluate the changes in the proteome that occur after OGT inhibition or knock-down, employing a reverse-phase protein array (RPPA). We identify transcriptional repressor - hairy and enhancer of split-1 (HES1) - as a mediator of the OGT inhibition response in the TNBC cells. Inhibition of OGT as well as the loss of HES1 results in potent cytotoxicity and apoptosis. The study raises a possibility of using OGT inhibition to potentiate DNA damage in the TNBC cells.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

