High proportions of bacteria are culturable across major biomes
- PMID: 30952994
- PMCID: PMC6775996
- DOI: 10.1038/s41396-019-0410-3
High proportions of bacteria are culturable across major biomes
Abstract
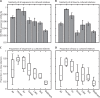
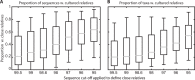
The paradigm that only 1% of microbes are culturable has had a profound impact on our understanding of microbial ecology and is still a major motivation for mostly using molecular tools to characterize microbial communities. However, this point is often expressed vaguely, suggesting that some scientists have different interpretations of the paradigm. In addition, there have been substantial advances in cultivation techniques suggesting that this paradigm may no longer be correct. To quantify bacterial culturability across six major biomes, I found that the median 16S rRNA similarity of bacteria to known cultured relatives was 97.3 ± 2.3% (s.d.). Furthermore, 52.0 ± 24% of sequences and 34.9 ± 23% of taxa (defined as >97% similar) had a closely related cultured relative. Thus, many cells and taxa across environments are culturable with known techniques, suggesting that the 1% paradigm is no longer correct.
Conflict of interest statement
The author declare that he has no conflict of interest.
Figures
Comment in
-
High proportions of bacteria and archaea across most biomes remain uncultured.ISME J. 2019 Dec;13(12):3126-3130. doi: 10.1038/s41396-019-0484-y. Epub 2019 Aug 6. ISME J. 2019. PMID: 31388130 Free PMC article.
-
The '1% culturability paradigm' needs to be carefully defined.ISME J. 2020 Jan;14(1):10-11. doi: 10.1038/s41396-019-0507-8. Epub 2019 Sep 24. ISME J. 2020. PMID: 31551529 Free PMC article. No abstract available.
References
-
- Brock TD. The study of microorganisms in situ: progress and problems. Symp Soc Gen Microbiol. 1987;41:1–17.

