Are innovation and new technologies in precision medicine paving a new era in patients centric care?
- PMID: 30953518
- PMCID: PMC6451233
- DOI: 10.1186/s12967-019-1864-9
Are innovation and new technologies in precision medicine paving a new era in patients centric care?
Abstract
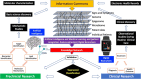
Healthcare is undergoing a transformation, and it is imperative to leverage new technologies to generate new data and support the advent of precision medicine (PM). Recent scientific breakthroughs and technological advancements have improved our understanding of disease pathogenesis and changed the way we diagnose and treat disease leading to more precise, predictable and powerful health care that is customized for the individual patient. Genetic, genomics, and epigenetic alterations appear to be contributing to different diseases. Deep clinical phenotyping, combined with advanced molecular phenotypic profiling, enables the construction of causal network models in which a genomic region is proposed to influence the levels of transcripts, proteins, and metabolites. Phenotypic analysis bears great importance to elucidat the pathophysiology of networks at the molecular and cellular level. Digital biomarkers (BMs) can have several applications beyond clinical trials in diagnostics-to identify patients affected by a disease or to guide treatment. Digital BMs present a big opportunity to measure clinical endpoints in a remote, objective and unbiased manner. However, the use of "omics" technologies and large sample sizes have generated massive amounts of data sets, and their analyses have become a major bottleneck requiring sophisticated computational and statistical methods. With the wealth of information for different diseases and its link to intrinsic biology, the challenge is now to turn the multi-parametric taxonomic classification of a disease into better clinical decision-making by more precisely defining a disease. As a result, the big data revolution has provided an opportunity to apply artificial intelligence (AI) and machine learning algorithms to this vast data set. The advancements in digital health opportunities have also arisen numerous questions and concerns on the future of healthcare practices in particular with what regards the reliability of AI diagnostic tools, the impact on clinical practice and vulnerability of algorithms. AI, machine learning algorithms, computational biology, and digital BMs will offer an opportunity to translate new data into actionable information thus, allowing earlier diagnosis and precise treatment options. A better understanding and cohesiveness of the different components of the knowledge network is a must to fully exploit the potential of it.
Keywords: Artificial intelligence; Autoimmune and inflammatory diseases; Biomarkers; Cancer; Deep phenotyping; Diabetes; Digital biomarkers; Epigenetics; Genetics; Genomics; Immuno-oncology; Machine learning; Modeling and simulation; Personalized medicine; Precision medicine; Proteomics; Transcriptomics; miRNAs; microRNAs.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Seyhan A, Carini C. Biomarkers for drug development: the time is now. Carini C, Menon S, Chang M, editors. Clinical and statistical considerations in personalized medicine. Chapman & Hall: CRC Press; 2014. p. 16–41.
-
- Seyhan AA. Biomarkers in drug discovery and development. Eur Biopharm Rev. 2010;1:19–25.
-
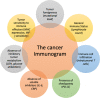
- Blank CU, Haanen JB, Ribas A, Schumacher TN. The “cancer immunogram”. Science. 2016;352:658–660. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

