Unbiased Pathogen Detection and Host Gene Profiling for Conjunctivitis
- PMID: 30953744
- PMCID: PMC6646074
- DOI: 10.1016/j.ophtha.2019.03.039
Unbiased Pathogen Detection and Host Gene Profiling for Conjunctivitis
Abstract
Purpose: The etiology of conjunctivitis is often misdiagnosed. An ideal diagnostic test would identify all possible infectious causes. In this study, we apply unbiased metagenomic RNA deep sequencing (MDS) to identify pathogens causing conjunctivitis.
Design: Molecular study of prospectively collected conjunctival swabs from patients with presumed infectious conjunctivitis.
Participants: Patients with presumed acute infectious conjunctivitis.
Methods: Conjunctival swabs were collected from patients presenting with acute conjunctivitis. Swabs were processed for MDS. Pathogens were identified using a rapid computational pipeline to analyze the nonhost sequences obtained from MDS. Differential gene expression analysis was performed to evaluate for host transcriptome signatures for infectious types. Clinical samples were deidentified, and laboratory personnel handling the samples and interpreting the data were masked.
Main outcome measures: Pathogens and differential transcripts identified by MDS.
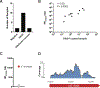
Results: Metagenomic RNA deep sequencing detected pathogens in 86% (12/14) of the patients tested. Swabs from 10 of 14 patients were positive for human adenovirus (HAdV) while swabs from 2 of 14 patients were positive for Vittaforma corneae (a parasitic fungal species of the microsporidia group). Samples positive for HAdV by RNA-seq were independently verified in a CLIA-certified laboratory. Pathogen-directed polymerase chain reaction confirmed the presence of V. corneae genome in the samples positive by RNA-seq. Local host transcriptome analysis identified 12 differentially expressed genes that provided distinct expression signatures for patients infected with HAdV compared with V. corneae.
Conclusions: Metagenomic RNA deep sequencing can reliably detect and quantify common and rare pathogens causing conjunctivitis, and identify strains. The unbiased nature of metagenomic RNA deep sequencing allowed an expanded scope of pathogen detection, including fungal species not commonly associated with acute conjunctivitis. In addition, the identification of infection type-specific local host transcriptome signatures may allow for pathogen detection even when the pathogen load is too low for direct identification.
Copyright © 2019 American Academy of Ophthalmology. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- O’Brien TP, Jeng BH, McDonald M, Raizman MB. Acute conjunctivitis: truth and misconceptions. Curr Med Res Opin 2009;25(8):1953–61. - PubMed
-
- Cheung D, Bremner J, Chan JT. Epidemic kerato-conjunctivitis--do outbreaks have to be epidemic? Eye (Lond) 2003;17(3):356–63. - PubMed
-
- Bharathi MJ, Murugan N, Kumar GR, et al. Vittaforma corneae keratitis in southern India: role of a novel duplex PCR. J Med Microbiol 2013;62(Pt 4):553–9. - PubMed
-
- Leveque N, Huguet P, Norder H, Chomel JJ. [Enteroviruses responsible for acute hemorrhagic conjunctivitis]. Med Mal Infect 2010;40(4):212–8. - PubMed
-
- Agashe R, Radhakrishnan N, Pradhan S, et al. Clinical and demographic study of microsporidial keratoconjunctivitis in South India: a 3-year study (2013–2015). Br J Ophthalmol 2017;101(10):1436–9. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

