Colonic Mucosal Transcriptomic Changes in Patients with Long-Duration Ulcerative Colitis Revealed Colitis-Associated Cancer Pathways
- PMID: 30954025
- PMCID: PMC6535502
- DOI: 10.1093/ecco-jcc/jjz002
Colonic Mucosal Transcriptomic Changes in Patients with Long-Duration Ulcerative Colitis Revealed Colitis-Associated Cancer Pathways
Abstract
Background and aims: Patients with ulcerative colitis [UC] with long disease duration have a higher risk of developing colitis-associated cancer [CAC] compared with patients with short-duration UC. The aim of this study was to identify transcriptomic differences associated with the duration of UC disease.
Methods: We conducted transcriptome profiling on 32 colonic biopsies [11 long-duration UC, ≥20 years; and 21 short-duration UC, ≤5 years] using Affymetrix Human Transcriptome Array 2.0. Differentially expressed genes [fold change > 1.5, p < 0.05] and alternative splicing events [splicing index > 1.5, p < 0.05] were determined using the Transcriptome Analysis Console. KOBAS 3.0 and DAVID 6.8 were used for KEGG and GO analysis. Selected genes from microarray analysis were validated using qPCR.
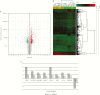
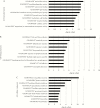
Results: There were 640 differentially expressed genes between both groups. The top ten upregulated genes were HMGCS2, UGT2A3 isoforms, B4GALNT2, MEP1B, GUCA2B, ADH1C, OTOP2, SLC9A3, and LYPD8; the top ten downregulated genes were PI3, DUOX2, VNN1, SLC6A14, GREM1, MMP1, CXCL1, TNIP3, TFF1, and LCN2. Among the 123 altered KEGG pathways, the most significant were metabolic pathways; fatty acid degradation; valine, leucine, and isoleucine degradation; the peroxisome proliferator-activated receptor signalling pathway; and bile secretion, which were previously linked with CAC. Analysis showed that 3560 genes exhibited differential alternative splicing between long- and short-duration UC. Among them, 374 were differentially expressed, underscoring the intrinsic relationship between altered gene expression and alternative splicing.
Conclusions: Long-duration UC patients have altered gene expressions, pathways, and alternative splicing events as compared with short-duration UC patients, and these could be further validated to improve our understanding of the pathogenesis of CAC.
Keywords: Inflammatory bowel disease; long duration; microarray analysis; transcriptome; ulcerative colitis.
© European Crohn’s and Colitis Organisation (ECCO) 2019.
Figures

References
-
- Ng SC, Shi HY, Hamidi N, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet 2018;390:2769–78. - PubMed
-
- Prideaux L, Kamm MA, De Cruz PP, Chan FK, Ng SC. Inflammatory bowel disease in Asia: a systematic review. J Gastroenterol Hepatol 2012;27:1266–80. - PubMed
-
- Hilmi I, Jaya F, Chua A, Heng WC, Singh H, Goh KL. A first study on the incidence and prevalence of IBD in Malaysia—results from the Kinta Valley IBD Epidemiology Study. J Crohns Colitis 2015;9:404–9. - PubMed
-
- Kim SC, Tonkonogy SL, Karrasch T, Jobin C, Sartor RB. Dual-association of gnotobiotic IL-10–/– mice with 2 nonpathogenic commensal bacteria induces aggressive pancolitis. Inflamm Bowel Dis 2007;13:1457–66. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

