Transfer of Functional Cargo in Exomeres
- PMID: 30956133
- PMCID: PMC6559347
- DOI: 10.1016/j.celrep.2019.01.009
Transfer of Functional Cargo in Exomeres
Abstract
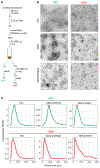
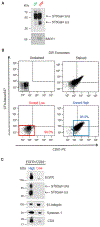
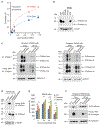
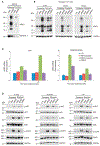
Exomeres are a recently discovered type of extracellular nanoparticle with no known biological function. Herein, we describe a simple ultracentrifugation-based method for separation of exomeres from exosomes. Exomeres are enriched in Argonaute 1-3 and amyloid precursor protein. We identify distinct functions of exomeres mediated by two of their cargo, the β-galactoside α2,6-sialyltransferase 1 (ST6Gal-I) that α2,6- sialylates N-glycans, and the EGFR ligand, amphiregulin (AREG). Functional ST6Gal-I in exomeres can be transferred to cells, resulting in hypersialylation of recipient cell-surface proteins including β1-integrin. AREG-containing exomeres elicit prolonged EGFR and downstream signaling in recipient cells, modulate EGFR trafficking in normal intestinal organoids, and dramatically enhance the growth of colonic tumor organoids. This study provides a simplified method of exomere isolation and demonstrates that exomeres contain and can transfer functional cargo. These findings underscore the heterogeneity of nanoparticles and should accelerate advances in determining the composition and biological functions of exomeres.
Keywords: Argonautes; EGFR; ST6Gal-I; amphiregulin; exomeres; exosomes; extracellular vesicles; fluorescence-activated vesicle sorting; organoids; β1-integrin.
Copyright © 2019 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures



References
-
- Barnard JA, Graves-Deal R, Pittelkow MR, DuBois R, Cook P, Ramsey GW, Bishop PR, Damstrup L, and Coffey RJ (1994). Auto- and cross-induction within the mammalian epidermal growth factor-related peptide family. J. Biol. Chem 269, 22817–22822. - PubMed
-
- Bligh EG, and Dyer WJ (1959). A rapid method oftotal lipid extraction and purification. Can. J. Biochem. Physiol 37, 911–917. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- K22 HL113039/HL/NHLBI NIH HHS/United States
- P50 CA095103/CA/NCI NIH HHS/United States
- R01 CA046413/CA/NCI NIH HHS/United States
- R35 CA197570/CA/NCI NIH HHS/United States
- P30 DK058404/DK/NIDDK NIH HHS/United States
- R01 HL127173/HL/NHLBI NIH HHS/United States
- U19 CA179514/CA/NCI NIH HHS/United States
- R01 HL128996/HL/NHLBI NIH HHS/United States
- UG3 CA241685/CA/NCI NIH HHS/United States
- R01 CA225177/CA/NCI NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- R01 CA163563/CA/NCI NIH HHS/United States
- P01 HL116263/HL/NHLBI NIH HHS/United States
- P50 CA236733/CA/NCI NIH HHS/United States
- T32 GM008111/GM/NIGMS NIH HHS/United States
- R01 GM111093/GM/NIGMS NIH HHS/United States
- U01 CA233581/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

