The PARK10 gene USP24 is a negative regulator of autophagy and ULK1 protein stability
- PMID: 30957634
- PMCID: PMC6984603
- DOI: 10.1080/15548627.2019.1598754
The PARK10 gene USP24 is a negative regulator of autophagy and ULK1 protein stability
Abstract
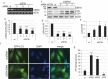
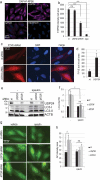
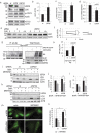
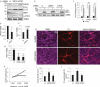
Recent studies indicate a causative relationship between defects in autophagy and dopaminergic neuron degeneration in Parkinson disease (PD). However, it is not fully understood how autophagy is regulated in the context of PD. Here we identify USP24 (ubiquitin specific peptidase 24), a gene located in the PARK10 (Parkinson disease 10 [susceptibility]) locus associated with late onset PD, as a novel negative regulator of autophagy. Our data indicate that USP24 regulates autophagy by affecting ubiquitination and stability of the ULK1 protein. Knockdown of USP24 in cell lines and in human induced-pluripotent stem cells (iPSC) differentiated into dopaminergic neurons resulted in elevated ULK1 protein levels and increased autophagy flux in a manner independent of MTORC1 but dependent on the class III phosphatidylinositol 3-kinase (PtdIns3K) activity. Surprisingly, USP24 knockdown also improved neurite extension and/or maintenance in aged iPSC-derived dopaminergic neurons. Furthermore, we observed elevated levels of USP24 in the substantia nigra of a subpopulation of idiopathic PD patients, suggesting that USP24 may negatively regulate autophagy in PD.Abbreviations: Bafilomycin/BafA: bafilomycin A1; DUB: deubiquitinating enzyme; iPSC: induced pluripotent stem cells; MTOR: mechanistic target of rapamycin kinase; MTORC1: MTOR complex 1; nt: non-targeting; PD: Parkinson disease; p-ATG13: phospho-ATG13; PtdIns3P: phosphatidylinositol 3-phosphate; RPS6: ribosomal protein S6; SNPs: single nucleotide polymorphisms; TH: tyrosine hydroxylase; USP24: ubiquitin specific peptidase 24.
Keywords: Autophagy; Parkinson disease; USP24; dopaminergic neurons; induced-pluripotent stem cells.
Figures


References
-
- Levine B, Klionsky DJ.. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004. April;6(4):463–477. PubMed PMID: 15068787. - PubMed
-
- Hara T, Nakamura K, Matsui M, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006. June 15;441(7095):885–889. PubMed PMID: 16625204. - PubMed
-
- Komatsu M, Waguri S, Chiba T, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006. June 15;441(7095):880–884. PubMed PMID: 16625205. - PubMed
-
- Lesage S, Brice A. Role of mendelian genes in “sporadic” Parkinson‘s disease. Parkinsonism Relat Disord. 2012. January;18 Suppl 1:S66–S70. PubMed PMID: 22166458; eng. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
