Characterizing Histone Post-translational Modification Alterations in Yeast Neurodegenerative Proteinopathy Models
- PMID: 30958470
- PMCID: PMC6543847
- DOI: 10.3791/59104
Characterizing Histone Post-translational Modification Alterations in Yeast Neurodegenerative Proteinopathy Models
Abstract
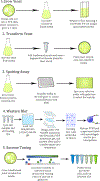
Neurodegenerative diseases, such as amyotrophic lateral sclerosis (ALS) and Parkinson's disease (PD), cause the loss of hundreds of thousands of lives each year. Effective treatment options able to halt disease progression are lacking. Despite the extensive sequencing efforts in large patient populations, the majority of ALS and PD cases remain unexplained by genetic mutations alone. Epigenetics mechanisms, such as the post-translational modification of histone proteins, may be involved in neurodegenerative disease etiology and progression and lead to new targets for pharmaceutical intervention. Mammalian in vivo and in vitro models of ALS and PD are costly and often require prolonged and laborious experimental protocols. Here, we outline a practical, fast, and cost-effective approach to determining genome-wide alterations in histone modification levels using Saccharomyces cerevisiae as a model system. This protocol allows for comprehensive investigations into epigenetic changes connected to neurodegenerative proteinopathies that corroborate previous findings in different model systems while significantly expanding our knowledge of the neurodegenerative disease epigenome.
Figures



References
-
- Kim HJ et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 495, 467, doi:10.1038/nature1192210.1038/nature11922https://www.nature.com/articles/nature11922-supplementary-informationhttps://www.nature.com/articles/nature11922-supplementary-information, (2013). - DOI - PMC - PubMed
-
- Poewe W et al. Parkinson disease. Nature Reviews Disease Primers. 3, 17013, doi:10.1038/nrdp.2017.1310.1038/nrdp.2017.13https://www.nature.com/articles/nrdp201713-supplementary-informationhttps://www.nature.com/articles/nrdp201713-supplementary-information, (2017). - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
