Machine Learning Based Real-Time Image-Guided Cell Sorting and Classification
- PMID: 30958640
- PMCID: PMC6520141
- DOI: 10.1002/cyto.a.23764
Machine Learning Based Real-Time Image-Guided Cell Sorting and Classification
Abstract
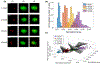
Cell classification based on phenotypical, spatial, and genetic information greatly advances our understanding of the physiology and pathology of biological systems. Technologies derived from next generation sequencing and fluorescent activated cell sorting are cornerstones for cell- and genomic-based assays supporting cell classification and mapping. However, there exists a deficiency in technology space to rapidly isolate cells based on high content image information. Fluorescence-activated cell sorting can only resolve cell-to-cell variation in fluorescence and optical scattering. Utilizing microfluidics, photonics, computation microscopy, real-time image processing and machine learning, we demonstrate an image-guided cell sorting and classification system possessing the high throughput of flow cytometer and high information content of microscopy. We demonstrate the utility of this technology in cell sorting based on (1) nuclear localization of glucocorticoid receptors, (2) particle binding to the cell membrane, and (3) DNA damage induced γ-H2AX foci. © 2019 International Society for Advancement of Cytometry.
Keywords: image guided cell sorting; imaging flow cytometry; machine learning; microfluidic.
© 2019 International Society for Advancement of Cytometry.
Figures




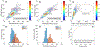
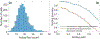
References
-
- Herzenberg LA, Parks D, Sahaf B, Perez O, Roederer M, & Herzenberg LA (2002). The history and future of the fluorescence activated cell sorter and flow cytometry: a view from Stanford. Clinical chemistry, 48(10), 1819–1827. - PubMed
-
- Nitta N, Sugimura T, Isozaki A, Mikami H, Hiraki K, Sakuma S, … & Fukuzawa H (2018). Intelligent image-activated cell sorting. Cell, 175(1), 266–276. - PubMed
-
- Juan G, Hernando E, & Cordon‐Cardo C (2002). Separation of live cells in different phases of the cell cycle for gene expression analysis. Cytometry Part A, 49(4), 170–175. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

