Single-cell RNA sequencing identifies inflammatory tissue T cells in eosinophilic esophagitis
- PMID: 30958799
- PMCID: PMC6486341
- DOI: 10.1172/JCI125917
Single-cell RNA sequencing identifies inflammatory tissue T cells in eosinophilic esophagitis
Abstract
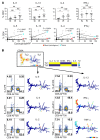
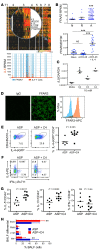
T cell heterogeneity is highly relevant to allergic disorders. We resolved the heterogeneity of human tissue CD3+ T cells during allergic inflammation, focusing on a tissue-specific allergic disease, eosinophilic esophagitis (EoE). We investigated 1088 single T cells derived from patients with a spectrum of disease activity. Eight disparate tissue T cell subtypes (designated T1-T8) were identified, with T7 and T8 enriched in the diseased tissue. The phenotypes of T7 and T8 resemble putative Treg (FOXP3+) and effector Th2-like (GATA3+) cells, respectively. Prodigious levels of IL-5 and IL-13 were confined to HPGDS+ CRTH2+IL-17RB+FFAR3+CD4+ T8 effector Th2 cells. EoE severity closely paralleled a lipid/fatty acid-induced activation node highlighted by the expression of the short-chain fatty acid receptor FFAR3. Ligands for FFAR3 induced Th2 cytokine production from human and murine T cells, including in an in vivo allergy model. Therefore, we elucidated the defining characteristics of tissue-residing CD3+ T cells in EoE, a specific enrichment of CD4+ Treg and effector Th2 cells, confinement of type 2 cytokine production to the CD4+ effector population, a highly likely role for FFAR3 in amplifying local Th2 responses in EoE, and a resource to further dissect tissue lymphocytes and allergic responses.
Keywords: Allergy; Immunology.
Conflict of interest statement
Figures






Comment in
-
Singling out Th2 cells in eosinophilic esophagitis.J Clin Invest. 2019 Apr 8;129(5):1830-1832. doi: 10.1172/JCI128479. eCollection 2019 Apr 8. J Clin Invest. 2019. PMID: 30958801 Free PMC article.
References
-
- Ziegler SF. Division of labour by CD4(+) T helper cells. Nat Rev Immunol. 2016;16(7):403. - PubMed
-
- Hirano I, et al. Dupilumab efficacy and safety in adult patients with active eosinophilic esophagitis: a randomized double-blind placebo-controlled phase 2 trial. Presented at the American College of Gastroenterology annual scientific meeting. Orlando, Florida. October 16, 2017.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

