Enhanced autophagic-lysosomal activity and increased BAG3-mediated selective macroautophagy as adaptive response of neuronal cells to chronic oxidative stress
- PMID: 30959460
- PMCID: PMC6454062
- DOI: 10.1016/j.redox.2019.101181
Enhanced autophagic-lysosomal activity and increased BAG3-mediated selective macroautophagy as adaptive response of neuronal cells to chronic oxidative stress
Abstract
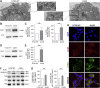
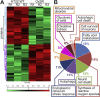
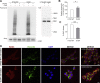
Oxidative stress and a disturbed cellular protein homeostasis (proteostasis) belong to the most important hallmarks of aging and of neurodegenerative disorders. The proteasomal and autophagic-lysosomal degradation pathways are key measures to maintain proteostasis. Here, we report that hippocampal cells selected for full adaptation and resistance to oxidative stress induced by hydrogen peroxide (oxidative stress-resistant cells, OxSR cells) showed a massive increase in the expression of components of the cellular autophagic-lysosomal network and a significantly higher overall autophagic activity. A comparative expression analysis revealed that distinct key regulators of autophagy are upregulated in OxSR cells. The observed adaptive autophagic response was found to be independent of the upstream autophagy regulator mTOR but is accompanied by a significant upregulation of further downstream components of the canonical autophagy network such as Beclin1, WIPI1 and the transmembrane ATG9 proteins. Interestingly, the expression of the HSP70 co-chaperone BAG3, mediator of BAG3-mediated selective macroautophagy and highly relevant for the clearance of aggregated proteins in cells, was found to be increased in OxSR cells that were consequently able to effectively overcome proteotoxic stress. Overexpression of BAG3 in oxidative stress-sensitive HT22 wildtype cells partly established the vesicular phenotype and the enhanced autophagic flux seen in OxSR cells suggesting that BAG3 takes over an important part in the adaptation process. A full proteome analysis demonstrated additional changes in the expression of mitochondrial proteins, metabolic enzymes and different pathway regulators in OxSR cells as consequence of the adaptation to oxidative stress in addition to autophagy-related proteins. Taken together, this analysis revealed a wide variety of pathways and players that act as adaptive response to chronic redox stress in neuronal cells.
Keywords: Adaptation; Autophagy; BAG3; Oxidative stress; Protein homeostasis.
Copyright © 2019. Published by Elsevier B.V.
Figures




References
-
- Hensley K., Hall N., Subramaniam R., Cole P., Harris M., Aksenov M., Aksenova M., Gabbita S.P., Wu J.F., Carney J.M. Brain regional correspondence between Alzheimer's disease histopathology and biomarkers of protein oxidation. J. Neurochem. 1995;65:2146–2156. - PubMed
-
- Di Domenico F., Tramutola A., Butterfield D.A. Role of 4-hydroxy-2-nonenal (HNE) in the pathogenesis of alzheimer disease and other selected age-related neurodegenerative disorders. Free Radic. Biol. Med. 2017;111:253–261. - PubMed
-
- Go´ mez-Isla T., Hollister R., West H., Mui S., Growdon J.H., Petersen C.R., Parisi E.J., Hyman B.T. Neuronl loss correlates with but exceeds neurofibrillary tangles in Alzheimer's disease. Am. Neurol. Associat. 1996;41:17–24. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

