Confirmation of Bioinformatics Predictions of the Structural Domains in Honeybee Silk
- PMID: 30960701
- PMCID: PMC6403662
- DOI: 10.3390/polym10070776
Confirmation of Bioinformatics Predictions of the Structural Domains in Honeybee Silk
Abstract
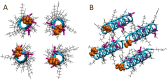
Honeybee larvae produce a silk made up of proteins in predominantly a coiled coil molecular structure. These proteins can be produced in recombinant systems, making them desirable templates for the design of advanced materials. However, the atomic level structure of these proteins is proving difficult to determine: firstly, because coiled coils are difficult to crystalize; and secondly, fibrous proteins crystalize as fibres rather than as discrete protein units. In this study, we synthesised peptides from the central structural domain, as well as the N- and C-terminal domains, of the honeybee silk. We used circular dichroism spectroscopy, infrared spectroscopy, and molecular dynamics to investigate the folding behaviour of the central domain peptides. We found that they folded as predicted by bioinformatics analysis, giving the protein engineer confidence in bioinformatics predictions to guide the design of new functionality into these protein templates. These results, along with the infrared structural analysis of the N- and C-terminal domain peptides and the comparison of peptide film properties with those of the full-length AmelF3 protein, provided significant insight into the structural elements required for honeybee silk protein to form into stable materials.
Keywords: bioinformatics protein folding prediction; cast film solubility; circular dichroism spectroscopy; coiled coil; infrared spectroscopy; molecular dynamics; protein design; protein engineering; protein materials; protein secondary structure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



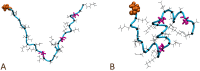
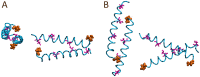



References
-
- Hepburn H.R., Kurstjens S.P. The combs of honeybees as composite materials. Apidologie. 1988;19:25–36. doi: 10.1051/apido:19880102. - DOI
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

