CRISPR/Cas9-mediated targeted mutagenesis of GmSPL9 genes alters plant architecture in soybean
- PMID: 30961525
- PMCID: PMC6454688
- DOI: 10.1186/s12870-019-1746-6
CRISPR/Cas9-mediated targeted mutagenesis of GmSPL9 genes alters plant architecture in soybean
Abstract
Background: The plant architecture has significant effects on grain yield of various crops, including soybean (Glycine max), but the knowledge on optimization of plant architecture in order to increase yield potential is still limited. Recently, CRISPR/Cas9 system has revolutionized genome editing, and has been widely utilized to edit the genomes of a diverse range of crop plants.
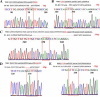
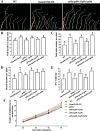
Results: In the present study, we employed the CRISPR/Cas9 system to mutate four genes encoding SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) transcription factors of the SPL9 family in soybean. These four GmSPL9 genes are negatively regulated by GmmiR156b, a target for the improvement of soybean plant architecture and yields. The soybean Williams 82 was transformed with the binary CRISPR/Cas9 plasmid, assembled with four sgRNA expression cassettes driven by the Arabidopsis thaliana U3 or U6 promoter, targeting different sites of these four SPL9 genes via Agrobacterium tumefaciens-mediated transformation. A 1-bp deletion was detected in one target site of the GmSPL9a and one target site of the GmSPL9b, respectively, by DNA sequencing analysis of two T0-generation plants. T2-generation spl9a and spl9b homozygous single mutants exhibited no obvious phenotype changes; but the T2 double homozygous mutant spl9a/spl9b possessed shorter plastochron length. In T4 generation, higher-order mutant plants carrying various combinations of mutations showed increased node number on the main stem and branch number, consequently increased total node number per plants at different levels. In addition, the expression levels of the examined GmSPL9 genes were higher in the spl9b-1 single mutant than wild-type plants, which might suggest a feedback regulation on the expression of the investigated GmSPL9 genes in soybean.
Conclusions: Our results showed that CRISPR/Cas9-mediated targeted mutagenesis of four GmSPL9 genes in different combinations altered plant architecture in soybean. The findings demonstrated that GmSPL9a, GmSPL9b, GmSPL9c and GmSPL9 function as redundant transcription factors in regulating plant architecture in soybean.
Keywords: CRISPR/Cas9; Plant architecture; Plastochron length; SPL; Soybean.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures



References
-
- Hartung RC, Specht JE, Williams JH. Modification of soybean plant architecture by genes for stem growth habit and maturity. Crop Sci. 1981;21:51–56. doi: 10.2135/cropsci1981.0011183X002100010015x. - DOI
-
- Huyghe C. Genetics and genetic modifications of plant architecture in grain legumes: a review. Agronomie. 1998;18:383–411. doi: 10.1051/agro:19980505. - DOI
-
- Bernard RL. Two genes affecting stem termination in soybean. Crop Sci. 1972;12:235–239. doi: 10.2135/cropsci1972.0011183X001200020028x. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

