Blood Microbiome Profile in CKD : A Pilot Study
- PMID: 30962186
- PMCID: PMC6500932
- DOI: 10.2215/CJN.12161018
Blood Microbiome Profile in CKD : A Pilot Study
Abstract
Background and objectives: The association between gut dysbiosis, high intestinal permeability, and endotoxemia-mediated inflammation is well established in CKD. However, changes in the circulating microbiome in patients with CKD have not been studied. In this pilot study, we compare the blood microbiome profile between patients with CKD and healthy controls using 16S ribosomal DNA sequencing.
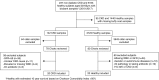
Design, setting, participants, & measurements: Blood bacterial DNA was studied in buffy coat samples quantitatively by 16S PCR and qualitatively by 16S targeted metagenomic sequencing using a molecular pipeline specifically optimized for blood samples in a cross-sectional study comparing 20 nondiabetic patients with CKD and 20 healthy controls.
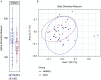
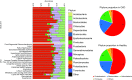
Results: There were 22 operational taxonomic units significantly different between the two groups. 16S metagenomic sequencing revealed a significant reduction in α diversity (Chao1 index) in the CKD group compared with healthy controls (127±18 versus 145±31; P=0.04). Proteobacteria phylum, Gammaproteobacteria class, and Enterobacteriaceae and Pseudomonadaceae families were more abundant in the CKD group compared with healthy controls. Median 16S ribosomal DNA levels did not significantly differ between CKD and healthy groups (117 versus 122 copies/ng DNA; P=0.38). GFR correlated inversely with the proportion of Proteobacteria (r=-0.54; P≤0.01).
Conclusions: Our pilot study demonstrates qualitative differences in the circulating microbiome profile with lower α diversity and significant taxonomic variations in the blood microbiome in patients with CKD compared with healthy controls.
Keywords: Blood microbiome; Cross-Sectional Studies; DNA, Bacterial; DNA, Ribosomal; Dysbiosis; Endotoxemia; Enterobacteriaceae; Metagenomics; Microbiota; Permeability; Pilot Projects; Polymerase Chain Reaction; Proteobacteria; Pseudomonadaceae; Renal Insufficiency, Chronic; Sequence Analysis, DNA; chronic kidney disease; glomerular filtration rate.
Copyright © 2019 by the American Society of Nephrology.
Figures


Comment in
-
Blood Microbiome in CKD: Should We Care?Clin J Am Soc Nephrol. 2019 May 7;14(5):648-649. doi: 10.2215/CJN.03420319. Epub 2019 Apr 8. Clin J Am Soc Nephrol. 2019. PMID: 30962188 Free PMC article. No abstract available.
References
-
- Hooper LV, Gordon JI: Commensal host-bacterial relationships in the gut. Science 292: 1115–1118, 2001 - PubMed
-
- Hooper LV, Bry L, Falk PG, Gordon JI: Host-microbial symbiosis in the mammalian intestine: Exploring an internal ecosystem. BioEssays 20: 336–343, 1998 - PubMed
-
- Wang B, Yao M, Lv L, Ling Z, Li L: The human microbiota in health and disease. Engineering 3: 71–82, 2017
-
- Evenepoel P, Poesen R, Meijers B: The gut-kidney axis. Pediatr Nephrol 32: 2005–2014, 2017 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

