Longitudinal profiling of human blood transcriptome in healthy and lupus pregnancy
- PMID: 30962246
- PMCID: PMC6504211
- DOI: 10.1084/jem.20190185
Longitudinal profiling of human blood transcriptome in healthy and lupus pregnancy
Abstract
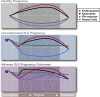
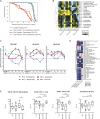
Systemic lupus erythematosus carries an increased risk of pregnancy complications, including preeclampsia and fetal adverse outcomes. To identify the underlying molecular mechanisms, we longitudinally profiled the blood transcriptome of 92 lupus patients and 43 healthy women during pregnancy and postpartum and performed multicolor flow cytometry in a subset of them. We also profiled 25 healthy women undergoing assisted reproductive technology to monitor transcriptional changes around embryo implantation. Sustained down-regulation of multiple immune signatures, including interferon and plasma cells, was observed during healthy pregnancy. These changes appeared early after embryo implantation and were mirrored in uncomplicated lupus pregnancies. Patients with preeclampsia displayed early up-regulation of neutrophil signatures that correlated with expansion of immature neutrophils. Lupus pregnancies with fetal complications carried the highest interferon and plasma cell signatures as well as activated CD4+ T cell counts. Thus, blood immunomonitoring reveals that both healthy and uncomplicated lupus pregnancies exhibit early and sustained transcriptional modulation of lupus-related signatures, and a lack thereof associates with adverse outcomes.
© 2019 Hong et al.
Figures






Comment in
-
When pregnancy tames the wolf.J Exp Med. 2019 May 6;216(5):1012-1013. doi: 10.1084/jem.20190378. Epub 2019 Apr 8. J Exp Med. 2019. PMID: 30962247 Free PMC article.
References
-
- ACOG Committee on Practice Bulletins–Obstetrics 2002. ACOG practice bulletin. Diagnosis and management of preeclampsia and eclampsia. Number 33, January 2002. Obstet. Gynecol. 99:159–167. - PubMed
-
- Allantaz F., Chaussabel D., Stichweh D., Bennett L., Allman W., Mejias A., Ardura M., Chung W., Smith E., Wise C., et al. . 2007. Blood leukocyte microarrays to diagnose systemic onset juvenile idiopathic arthritis and follow the response to IL-1 blockade [published correction appears in J Exp Med. 2009;206(10):2299]. J. Exp. Med. 204:2131–2144. 10.1084/jem.20070070 - DOI - PMC - PubMed
-
- Andrade D., Kim M., Blanco L.P., Karumanchi S.A., Koo G.C., Redecha P., Kirou K., Alvarez A.M., Mulla M.J., Crow M.K., et al. . 2015. Interferon-α and angiogenic dysregulation in pregnant lupus patients who develop preeclampsia. Arthritis Rheumatol. 67:977–987. 10.1002/art.39029 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

