Unraveling Hidden Components of the Chloroplast Envelope Proteome: Opportunities and Limits of Better MS Sensitivity
- PMID: 30962257
- PMCID: PMC6601204
- DOI: 10.1074/mcp.RA118.000988
Unraveling Hidden Components of the Chloroplast Envelope Proteome: Opportunities and Limits of Better MS Sensitivity
Abstract
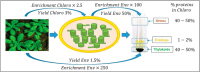
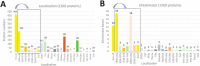
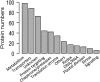
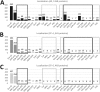
The chloroplast is a major plant cell organelle that fulfills essential metabolic and biosynthetic functions. Located at the interface between the chloroplast and other cell compartments, the chloroplast envelope system is a strategic barrier controlling the exchange of ions, metabolites and proteins, thus regulating essential metabolic functions (synthesis of hormones precursors, amino acids, pigments, sugars, vitamins, lipids, nucleotides etc.) of the plant cell. However, unraveling the contents of the chloroplast envelope proteome remains a difficult challenge; many proteins constituting this functional double membrane system remain to be identified. Indeed, the envelope contains only 1% of the chloroplast proteins (i.e. 0.4% of the whole cell proteome). In other words, most envelope proteins are so rare at the cell, chloroplast, or even envelope level, that they remained undetectable using targeted MS studies. Cross-contamination of chloroplast subcompartments by each other and by other cell compartments during cell fractionation, impedes accurate localization of many envelope proteins. The aim of the present study was to take advantage of technologically improved MS sensitivity to better define the proteome of the chloroplast envelope (differentiate genuine envelope proteins from contaminants). This MS-based analysis relied on an enrichment factor that was calculated for each protein identified in purified envelope fractions as compared with the value obtained for the same protein in crude cell extracts. Using this approach, a total of 1269 proteins were detected in purified envelope fractions, of which, 462 could be assigned an envelope localization by combining MS-based spectral count analyses with manual annotation using data from the literature and prediction tools. Many of such proteins being previously unknown envelope components, these data constitute a new resource of significant value to the broader plant science community aiming to define principles and molecular mechanisms controlling fundamental aspects of plastid biogenesis and functions.
Keywords: Cell fractionation*; Cellular organelles*; Chloroplast; Chloroplast envelope; Plant Biology*; Subcellular Separation; Subcellular analysis.
© 2019 Bouchnak et al.
Figures






Similar articles
-
AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins.Mol Cell Proteomics. 2010 Jun;9(6):1063-84. doi: 10.1074/mcp.M900325-MCP200. Epub 2010 Jan 10. Mol Cell Proteomics. 2010. PMID: 20061580 Free PMC article.
-
Integral membrane proteins of the chloroplast envelope: identification and subcellular localization of new transporters.Proc Natl Acad Sci U S A. 2002 Aug 20;99(17):11487-92. doi: 10.1073/pnas.172390399. Epub 2002 Aug 12. Proc Natl Acad Sci U S A. 2002. PMID: 12177442 Free PMC article.
-
The distinct functional roles of the inner and outer chloroplast envelope of Pea (Pisum sativum) as revealed by proteomic approaches.J Proteome Res. 2014 Jun 6;13(6):2941-53. doi: 10.1021/pr500106s. Epub 2014 May 13. J Proteome Res. 2014. PMID: 24792535
-
Biogenesis of chloroplast outer envelope membrane proteins.Plant Cell Rep. 2019 Jul;38(7):783-792. doi: 10.1007/s00299-019-02381-6. Epub 2019 Jan 22. Plant Cell Rep. 2019. PMID: 30671649 Review.
-
New Insights into the Chloroplast Outer Membrane Proteome and Associated Targeting Pathways.Int J Mol Sci. 2022 Jan 29;23(3):1571. doi: 10.3390/ijms23031571. Int J Mol Sci. 2022. PMID: 35163495 Free PMC article. Review.
Cited by
-
Ubiquitin-based pathway acts inside chloroplasts to regulate photosynthesis.Sci Adv. 2022 Nov 18;8(46):eabq7352. doi: 10.1126/sciadv.abq7352. Epub 2022 Nov 16. Sci Adv. 2022. PMID: 36383657 Free PMC article.
-
Mixotrophic growth of the extremophile Galdieria sulphuraria reveals the flexibility of its carbon assimilation metabolism.New Phytol. 2021 Jul;231(1):326-338. doi: 10.1111/nph.17359. Epub 2021 May 1. New Phytol. 2021. PMID: 33764540 Free PMC article.
-
Calmodulin is involved in the dual subcellular location of two chloroplast proteins.J Biol Chem. 2019 Nov 15;294(46):17543-17554. doi: 10.1074/jbc.RA119.010846. Epub 2019 Oct 2. J Biol Chem. 2019. PMID: 31578278 Free PMC article.
-
Chloroplast pH Homeostasis for the Regulation of Photosynthesis.Front Plant Sci. 2022 May 25;13:919896. doi: 10.3389/fpls.2022.919896. eCollection 2022. Front Plant Sci. 2022. PMID: 35693183 Free PMC article. Review.
-
Genome communication in plants mediated by organelle-n-ucleus-located proteins.Philos Trans R Soc Lond B Biol Sci. 2020 Jun 22;375(1801):20190397. doi: 10.1098/rstb.2019.0397. Epub 2020 May 4. Philos Trans R Soc Lond B Biol Sci. 2020. PMID: 32362260 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

