Carbapenem-Susceptible OXA-23-Producing Proteus mirabilis in the French Community
- PMID: 30962345
- PMCID: PMC6535531
- DOI: 10.1128/AAC.00191-19
Carbapenem-Susceptible OXA-23-Producing Proteus mirabilis in the French Community
Abstract
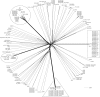
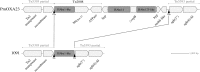
Nineteen Proteus mirabilis isolates producing the carbapenemase OXA-23 were recovered over a 2-year period in 19 French hospitalized patients, of whom 12 had community onset infections. The isolates exhibited a slightly reduced susceptibility to carbapenems. Whole-genome analysis revealed that all 19 isolates formed a cluster compared to 149 other P. mirabilis isolates. Because of its susceptibility to carbapenems, this clone may be misidentified as a penicillinase producer while it constitutes a reservoir of the OXA-23-encoding gene in the community.
Keywords: OXA-23; Proteus mirabilis; carbapenemase; clonality; spread.
Copyright © 2019 American Society for Microbiology.
Figures
References
-
- Paul D, Ingti B, Bhattacharjee D, Maurya AP, Dhar D, Chakravarty A, Bhattacharjee A. 2017. An unusual occurrence of plasmid-mediated blaOXA-23 carbapenemase in clinical isolates of Escherichia coli from India. Int J Antimicrob Agents 49:642–645. doi: 10.1016/j.ijantimicag.2017.01.012. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

