ϕSa3mw Prophage as a Molecular Regulatory Switch of Staphylococcus aureus β-Toxin Production
- PMID: 30962356
- PMCID: PMC6597384
- DOI: 10.1128/JB.00766-18
ϕSa3mw Prophage as a Molecular Regulatory Switch of Staphylococcus aureus β-Toxin Production
Abstract
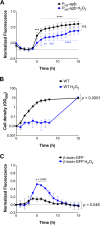
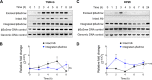
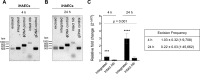
Phage regulatory switches (phage-RSs) are a newly described form of active lysogeny where prophages function as regulatory mechanisms for expression of chromosomal bacterial genes. In Staphylococcus aureus, ϕSa3int is a widely distributed family of prophages that integrate into the β-toxin structural gene hlb, effectively inactivating it. However, β-toxin-producing strains often arise during infections and are more virulent in experimental infective endocarditis and pneumonia infections. We present evidence that in S. aureus MW2, ϕSa3mw excision is temporally and differentially responsive to growth conditions relevant to S. aureus pathogenesis. PCR analyses of ϕSa3mw (integrated and excised) and of intact hlb showed that ϕSa3mw preferentially excises in response to hydrogen peroxide-induced oxidative stress and during biofilm growth. ϕSa3mw remains as a prophage when in contact with human aortic endothelial cells in culture. A criterion for a prophage to be considered a phage-RS is the inability to lyse host cells. MW2 grown under phage-inducing conditions did not release infectious phage particles by plaque assay or transmission electron microscopy, indicating that ϕSa3mw does not carry out a productive lytic cycle. These studies highlight a dynamic, and perhaps more sophisticated, S. aureus-prophage interaction where ϕSa3int prophages provide a novel regulatory mechanism for the conditional expression of virulence factors.IMPORTANCE β-Toxin is a sphingomyelinase hemolysin that significantly contributes to Staphylococcus aureus pathogenesis. In most S. aureus isolates the prophage ϕSa3int inserts into the β-toxin gene hlb, inactivating it, but human and experimental infections give rise to β-toxin-producing variants. However, it remained to be established whether ϕSa3mw excises in response to specific environmental cues, restoring the β-toxin gene sequence. This is not only of fundamental interest but also critical when designing intervention strategies and therapeutics. We provide evidence that ϕSa3mw actively excises, allowing the conditional expression of β-toxin. ϕSa3int prophages may play a novel and largely uncharacterized role in S. aureus pathogenesis as molecular regulatory switches that promote bacterial fitness and adaptation to the challenges presented by the mammalian host.
Keywords: PR-switch; Sa3int; Staphylococcus aureus; beta toxin; biofilm; infective endocarditis; oxidative stress; phage regulatory switch.
Copyright © 2019 Tran et al.
Figures


Similar articles
-
The Role of hlb-Converting Bacteriophages in Staphylococcus aureus Host Adaption.Microb Physiol. 2021;31(2):109-122. doi: 10.1159/000516645. Epub 2021 Jun 14. Microb Physiol. 2021. PMID: 34126612 Review.
-
Staphylococcus aureus β-toxin production is common in strains with the β-toxin gene inactivated by bacteriophage.J Infect Dis. 2014 Sep 1;210(5):784-92. doi: 10.1093/infdis/jiu146. Epub 2014 Mar 11. J Infect Dis. 2014. PMID: 24620023 Free PMC article.
-
Prophages encoding human immune evasion cluster genes are enriched in Staphylococcus aureus isolated from chronic rhinosinusitis patients with nasal polyps.Microb Genom. 2021 Dec;7(12):000726. doi: 10.1099/mgen.0.000726. Microb Genom. 2021. PMID: 34907894 Free PMC article.
-
Analysis of prophages harbored by the human-adapted subpopulation of Staphylococcus aureus CC398.Infect Genet Evol. 2013 Aug;18:299-308. doi: 10.1016/j.meegid.2013.06.009. Epub 2013 Jun 14. Infect Genet Evol. 2013. PMID: 23770143
-
Cryptic prophages as targets for drug development.Drug Resist Updat. 2016 Jul;27:30-8. doi: 10.1016/j.drup.2016.06.001. Epub 2016 Jun 6. Drug Resist Updat. 2016. PMID: 27449596 Review.
Cited by
-
Comparative Genomic Reveals Clonal Heterogeneity in Persistent Staphylococcus aureus Infection.Front Cell Infect Microbiol. 2022 Feb 21;12:817841. doi: 10.3389/fcimb.2022.817841. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 35265532 Free PMC article.
-
The Current Landscape of Phage-Antibiotic Synergistic (PAS) Interactions.Antibiotics (Basel). 2025 May 27;14(6):545. doi: 10.3390/antibiotics14060545. Antibiotics (Basel). 2025. PMID: 40558135 Free PMC article. Review.
-
CcpA Regulates Staphylococcus aureus Biofilm Formation through Direct Repression of Staphylokinase Expression.Antibiotics (Basel). 2022 Oct 17;11(10):1426. doi: 10.3390/antibiotics11101426. Antibiotics (Basel). 2022. PMID: 36290085 Free PMC article.
-
Staphylococcus aureus in Agriculture: Lessons in Evolution from a Multispecies Pathogen.Clin Microbiol Rev. 2021 Feb 10;34(2):e00182-20. doi: 10.1128/CMR.00182-20. Print 2021 Mar 17. Clin Microbiol Rev. 2021. PMID: 33568553 Free PMC article. Review.
-
In Silico Genome-Scale Analysis of Molecular Mechanisms Contributing to the Development of a Persistent Infection with Methicillin-Resistant Staphylococcus aureus (MRSA) ST239.Int J Mol Sci. 2022 Dec 16;23(24):16086. doi: 10.3390/ijms232416086. Int J Mol Sci. 2022. PMID: 36555727 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

