Ebolavirus polymerase uses an unconventional genome replication mechanism
- PMID: 30962389
- PMCID: PMC6486738
- DOI: 10.1073/pnas.1815745116
Ebolavirus polymerase uses an unconventional genome replication mechanism
Abstract
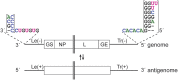
Most nonsegmented negative strand (NNS) RNA virus genomes have complementary 3' and 5' terminal nucleotides because the promoters at the 3' ends of the genomes and antigenomes are almost identical to each other. However, according to published sequences, both ends of ebolavirus genomes show a high degree of variability, and the 3' and 5' terminal nucleotides are not complementary. If correct, this would distinguish the ebolaviruses from other NNS RNA viruses. Therefore, we investigated the terminal genomic and antigenomic nucleotides of three different ebolavirus species, Ebola (EBOV), Sudan, and Reston viruses. Whereas the 5' ends of ebolavirus RNAs are highly conserved with the sequence ACAGG-5', the 3' termini are variable and are typically 3'-GCCUGU, ACCUGU, or CCUGU. A small fraction of analyzed RNAs had extended 3' ends. The majority of 3' terminal sequences are consistent with a mechanism of nucleotide addition by hairpin formation and back-priming. Using single-round replicating EBOV minigenomes, we investigated the effect of the 3' terminal nucleotide on viral replication and found that the EBOV polymerase initiates replication opposite the 3'-CCUGU motif regardless of the identity of the 3' terminal nucleotide(s) and of the position of this motif relative to the 3' end. Deletion or mutation of the first residue of the 3'-CCUGU motif completely abolished replication initiation, suggesting a crucial role of this nucleotide in directing initiation. Together, our data show that ebolaviruses have evolved a unique replication strategy among NNS RNA viruses resulting in 3' overhangs. This could be a mechanism to avoid antiviral recognition.
Keywords: Ebola virus genome ends; Ebola virus replication; ebolavirus replication initiation; nonsegmented negative strand RNA virus replication; variable 3′ genome ends.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Comment in
-
Ebola Virus Replication Stands Out.Trends Microbiol. 2019 Jul;27(7):565-566. doi: 10.1016/j.tim.2019.05.004. Epub 2019 May 30. Trends Microbiol. 2019. PMID: 31155428 Free PMC article.
References
-
- Whelan SP, Barr JN, Wertz GW. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr Top Microbiol Immunol. 2004;283:61–119. - PubMed
-
- Kiley MP, Wilusz J, McCormick JB, Keene JD. Conservation of the 3′ terminal nucleotide sequences of Ebola and Marburg virus. Virology. 1986;149:251–254. - PubMed
-
- Volchkov VE, et al. Characterization of the L gene and 5′ trailer region of Ebola virus. J Gen Virol. 1999;80:355–362. - PubMed
-
- Kolesnikova L, Nanbo A, Becker S, Kawaoka Y. Inside the cell: Assembly of filoviruses. Curr Top Microbiol Immunol. 2017;411:353–380. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

