Spaceflight-induced alternative splicing during seedling development in Arabidopsis thaliana
- PMID: 30963109
- PMCID: PMC6447593
- DOI: 10.1038/s41526-019-0070-7
Spaceflight-induced alternative splicing during seedling development in Arabidopsis thaliana
Abstract
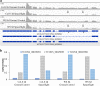
Plants grown in spaceflight experience novel environmental signals, including those associated with microgravity and ionizing radiation. Spaceflight triggers a response involving transcriptional re-programming and altered cell morphology, though many aspects of this response remain uncharacterized. We analyzed the spaceflight-induced transcriptome with a focus on genes that undergo alternative splicing to examine differential splicing associated with spaceflight-an unstudied characteristic of the molecular response to spaceflight exposure. RNA sequence data obtained during the APEX03 spaceflight experiment that was collected from two Arabidopsis thaliana ecotypes at two seedling stages grown onboard the International Space Station, or as ground controls at Kennedy Space Center, were re-examined to detect alternative splicing differences induced by spaceflight. Presence/absence variation analysis was used to identify putative expression-level differences in alternatively spliced isoforms between spaceflight and ground controls and was followed by analysis of significant differential alternative splicing. This study provides the first evidence of a role for alternative splicing in the molecular processes of physiological adaptation to the spaceflight environment.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
LinkOut - more resources
Full Text Sources

