Atypical URA5 gene restriction fragment length polymorphism banding profile in Cryptococcus neoformans strains
- PMID: 30963417
- PMCID: PMC6861435
- DOI: 10.1007/s12223-019-00699-y
Atypical URA5 gene restriction fragment length polymorphism banding profile in Cryptococcus neoformans strains
Abstract
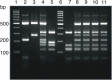
URA5-RFLP is one of the most widely used genotyping methods relating to Cryptococcus neoformans and C. gattii consensus genotype nomenclature. In order to identify a molecular type, this method uses a visual comparison of digested PCR products of tested and reference strains, therefore any anomaly in RFLP patterns of studied isolates makes recognition difficult or impossible. This report describes a strain of VNIV type showing an atypical URA5-RFLP pattern as well as a group of AD hybrids displaying the same anomaly. The atypical RFLP pattern is the result of a point mutation and emergence of a new restriction site. Emergence of the allele presenting a new banding pattern may lead to misidentification using the URA5-RFLP technique; the results of this study as well as the literature data may suggest the spread of the allele in the environment.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Cogliati M, Zani A, Rickerts V, McCormick I, Desnos-Ollivier M, Velegraki A, Escandon P, Ichikawa T, Ikeda R, Bienvenu AL, Tintelnot K, Tore O, Akcaglar S, Lockhart S, Tortorano AM, Varma A. Multilocus sequence typing analysis reveals that Cryptococcus neoformans var. neoformans is a recombinant population. Fungal Genet Biol. 2016;87:22–29. doi: 10.1016/j.fgb.2016.01.003. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

