BBX16, a B-box protein, positively regulates light-induced anthocyanin accumulation by activating MYB10 in red pear
- PMID: 30963689
- PMCID: PMC6737026
- DOI: 10.1111/pbi.13114
BBX16, a B-box protein, positively regulates light-induced anthocyanin accumulation by activating MYB10 in red pear
Abstract
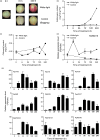
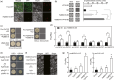
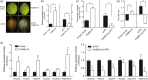
The red coloration of pear (Pyrus pyrifolia) results from anthocyanin accumulation in the fruit peel. Light is required for anthocyanin biosynthesis in pear. A pear homolog of Arabidopsis thaliana BBX22, PpBBX16, was differentially expressed after fruits were removed from bags and may be involved in anthocyanin biosynthesis. Here, the expression and function of PpBBX16 were analysed. PpBBX16's expression was highly induced by white-light irradiation, as was anthocyanin accumulation. PpBBX16's ectopic expression in Arabidopsis increased anthocyanin biosynthesis in the hypocotyls and tops of flower stalks. PpBBX16 was localized in the nucleus and showed trans-activity in yeast cells. Although PpBBX16 could not directly bind to the promoter of PpMYB10 or PpCHS in yeast one-hybrid assays, the complex of PpBBX16/PpHY5 strongly trans-activated anthocyanin pathway genes in tobacco. PpBBX16's overexpression in pear calli enhanced the red coloration during light treatments. Additionally, PpBBX16's transient overexpression in pear peel increased anthocyanin accumulation, while virus-induced gene silencing of PpBBX16 decreased anthocyanin accumulation. The expression patterns of pear BBX family members were analysed, and six additional BBX genes, which were differentially expressed during light-induced anthocyanin biosynthesis, were identified. Thus, PpBBX16 is a positive regulator of light-induced anthocyanin accumulation, but it could not directly induce the expression of the anthocyanin biosynthesis-related genes by itself but needed PpHY5 to gain full function. Our work uncovered regulatory modes for PpBBX16 and suggested the potential functions of other pear BBX genes in the regulation of anthocyanin accumulation, thereby providing target genes for further studies on anthocyanin biosynthesis.
Keywords: BBX16; MYB10; anthocyanin accumulation; light; pear.
© 2019 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
-
- Allan, A.C. and Espley, R.V. (2018) MYBs drive novel consumer traits in fruits and vegetables. Trends Plant Sci. 23, 693–705. - PubMed
-
- Almada, R. , Cabrera, N. , Casaretto, J.A. , RuizLara, S. and González, V.E. (2009) VvCO and VvCOL1, two CONSTANS homologous genes, are regulated during flower induction and dormancy in grapevine buds. Plant Cell Rep. 28, 1193–1203. - PubMed
-
- Bai, S.L. , Saito, T. , Honda, C. , Hatsuyama, Y. , Ito, A. and Moriguchi, T. (2014) An apple B‐box protein, MdCOL11, is involved in UV‐B‐ and temperature‐induced anthocyanin biosynthesis. Planta, 240, 1051–1062. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

