Estimating the total genome length of a metagenomic sample using k-mers
- PMID: 30967110
- PMCID: PMC6456951
- DOI: 10.1186/s12864-019-5467-x
Estimating the total genome length of a metagenomic sample using k-mers
Abstract
Background: Metagenomic sequencing is a powerful technology for studying the mixture of microbes or the microbiomes on human and in the environment. One basic task of analyzing metagenomic data is to identify the component genomes in the community. This task is challenging due to the complexity of microbiome composition, limited availability of known reference genomes, and usually insufficient sequencing coverage.
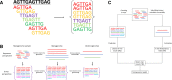
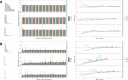
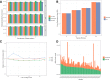
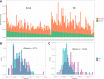
Results: As an initial step toward understanding the complete composition of a metagenomic sample, we studied the problem of estimating the total length of all distinct component genomes in a metagenomic sample. We showed that this problem can be solved by estimating the total number of distinct k-mers in all the metagenomic sequencing data. We proposed a method for this estimation based on the sequencing coverage distribution of observed k-mers, and introduced a k-mer redundancy index (KRI) to fill in the gap between the count of distinct k-mers and the total genome length. We showed the effectiveness of the proposed method on a set of carefully designed simulation data corresponding to multiple situations of true metagenomic data. Results on real data indicate that the uncaptured genomic information can vary dramatically across metagenomic samples, with the potential to mislead downstream analyses.
Conclusions: We proposed the question of how long the total genome length of all different species in a microbial community is and introduced a method to answer it.
Keywords: Distinct k-mers; Genome length; Metagenomics; Sequencing coverage.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- Cui H, Li Y, Zhang X. An overview of major metagenomic studies on human microbiomes in health and disease. Quant Biol. 2016;4(3):192–206. doi: 10.1007/s40484-016-0078-x. - DOI
-
- Zhang X, Liu S, Cui H, Chen T. Reading the underlying information from massive metagenomic sequencing data. Proc IEEE. 2017;105(3):459–73.
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

