Gene correction for SCID-X1 in long-term hematopoietic stem cells
- PMID: 30967552
- PMCID: PMC6456568
- DOI: 10.1038/s41467-019-09614-y
Gene correction for SCID-X1 in long-term hematopoietic stem cells
Erratum in
-
Author Correction: Gene correction for SCID-X1 in long-term hematopoietic stem cells.Nat Commun. 2019 Apr 26;10(1):2021. doi: 10.1038/s41467-019-10080-9. Nat Commun. 2019. PMID: 31028274 Free PMC article.
-
Author Correction: Gene correction for SCID-X1 in long-term hematopoietic stem cells.Nat Commun. 2019 Dec 4;10(1):5624. doi: 10.1038/s41467-019-13620-5. Nat Commun. 2019. PMID: 31796738 Free PMC article.
Abstract
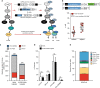
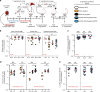
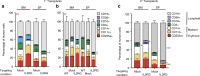
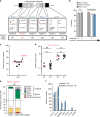
Gene correction in human long-term hematopoietic stem cells (LT-HSCs) could be an effective therapy for monogenic diseases of the blood and immune system. Here we describe an approach for X-linked sSevere cCombined iImmunodeficiency (SCID-X1) using targeted integration of a cDNA into the endogenous start codon to functionally correct disease-causing mutations throughout the gene. Using a CRISPR-Cas9/AAV6 based strategy, we achieve up to 20% targeted integration frequencies in LT-HSCs. As measures of the lack of toxicity we observe no evidence of abnormal hematopoiesis following transplantation and no evidence of off-target mutations using a high-fidelity Cas9 as a ribonucleoprotein complex. We achieve high levels of targeting frequencies (median 45%) in CD34+ HSPCs from six SCID-X1 patients and demonstrate rescue of lymphopoietic defect in a patient derived HSPC population in vitro and in vivo. In sum, our study provides specificity, toxicity and efficacy data supportive of clinical development of genome editing to treat SCID-Xl.
Conflict of interest statement
M.H.P. serves on the SAB for CRISPR Tx and Allogene Tx. Neither company had input into the design, execution, data analysis, or publication of the work presented in this manuscript. The remaining authors declare no competing interests.
Figures


Similar articles
-
Targeted genome editing in human repopulating haematopoietic stem cells.Nature. 2014 Jun 12;510(7504):235-240. doi: 10.1038/nature13420. Epub 2014 May 28. Nature. 2014. PMID: 24870228 Free PMC article.
-
Restoring T and B cell generation in X-linked severe combined immunodeficiency mice through hematopoietic stem cells adenine base editing.Mol Ther. 2024 Jun 5;32(6):1658-1671. doi: 10.1016/j.ymthe.2024.03.028. Epub 2024 Mar 26. Mol Ther. 2024. PMID: 38532630 Free PMC article.
-
Genome editing in human hematopoietic stem and progenitor cells via CRISPR-Cas9-mediated homology-independent targeted integration.Mol Ther. 2021 Apr 7;29(4):1611-1624. doi: 10.1016/j.ymthe.2020.12.010. Epub 2020 Dec 10. Mol Ther. 2021. PMID: 33309880 Free PMC article.
-
Gene therapy strategies for RAG1 deficiency: Challenges and breakthroughs.Immunol Lett. 2024 Dec;270:106931. doi: 10.1016/j.imlet.2024.106931. Epub 2024 Sep 18. Immunol Lett. 2024. PMID: 39303994 Review.
-
Gene therapy using haematopoietic stem and progenitor cells.Nat Rev Genet. 2021 Apr;22(4):216-234. doi: 10.1038/s41576-020-00298-5. Epub 2020 Dec 10. Nat Rev Genet. 2021. PMID: 33303992 Review.
Cited by
-
Answered and Unanswered Questions in Early-Stage Viral Vector Transduction Biology and Innate Primary Cell Toxicity for Ex-Vivo Gene Editing.Front Immunol. 2021 May 28;12:660302. doi: 10.3389/fimmu.2021.660302. eCollection 2021. Front Immunol. 2021. PMID: 34122418 Free PMC article. Review.
-
Genetically corrected RAG2-SCID human hematopoietic stem cells restore V(D)J-recombinase and rescue lymphoid deficiency.Blood Adv. 2024 Apr 9;8(7):1820-1833. doi: 10.1182/bloodadvances.2023011766. Blood Adv. 2024. PMID: 38096800 Free PMC article.
-
Correcting inborn errors of immunity: From viral mediated gene addition to gene editing.Semin Immunol. 2023 Mar;66:101731. doi: 10.1016/j.smim.2023.101731. Epub 2023 Feb 28. Semin Immunol. 2023. PMID: 36863140 Free PMC article. Review.
-
Advances in gene therapy for inborn errors of immunity.Curr Opin Allergy Clin Immunol. 2023 Dec 1;23(6):467-477. doi: 10.1097/ACI.0000000000000952. Epub 2023 Oct 13. Curr Opin Allergy Clin Immunol. 2023. PMID: 37846903 Free PMC article. Review.
-
Current approaches and potential challenges in the delivery of gene editing cargos into hematopoietic stem and progenitor cells.Front Genome Ed. 2023 Sep 15;5:1148693. doi: 10.3389/fgeed.2023.1148693. eCollection 2023. Front Genome Ed. 2023. PMID: 37780116 Free PMC article. Review.
References
-
- Stephan V, et al. Atypical X-linked severe combined immunodeficiency due to possible spontaneous reversion of the genetic defect in T cells. N. Engl. J. Med. 1996;335:1563–1567. - PubMed
-
- Hacein-Bey-Abina S, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. - PubMed
-
- Woods NB, Bottero V, Schmidt M, von Kalle C, Verma IM. Gene therapy: therapeutic gene causing lymphoma. Nature. 2006;440:1123. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

