Systems Biology Reveals NR2F6 and TGFB1 as Key Regulators of Feed Efficiency in Beef Cattle
- PMID: 30967894
- PMCID: PMC6439317
- DOI: 10.3389/fgene.2019.00230
Systems Biology Reveals NR2F6 and TGFB1 as Key Regulators of Feed Efficiency in Beef Cattle
Abstract
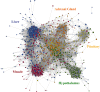
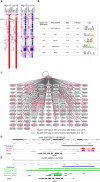
Systems biology approaches are used as strategy to uncover tissue-specific perturbations and regulatory genes related to complex phenotypes. We applied this approach to study feed efficiency (FE) in beef cattle, an important trait both economically and environmentally. Poly-A selected RNA of five tissues (adrenal gland, hypothalamus, liver, skeletal muscle and pituitary) of eighteen young bulls, selected for high and low FE, were sequenced (Illumina HiSeq 2500, 100 bp, pared-end). From the 17,354 expressed genes considering all tissues, 1,335 were prioritized by five selection categories (differentially expressed, harboring SNPs associated with FE, tissue-specific, secreted in plasma and key regulators) and used for network construction. NR2F6 and TGFB1 were identified and validated by motif discovery as key regulators of hepatic inflammatory response and muscle tissue development, respectively, two biological processes demonstrated to be associated with FE. Moreover, we indicated potential biomarkers of FE, which are related to hormonal control of metabolism and sexual maturity. By using robust methodologies and validation strategies, we confirmed the main biological processes related to FE in Bos indicus and indicated candidate genes as regulators or biomarkers of superior animals.
Keywords: Bos indicus; Nellore (Zebu); feed efficiency; inflammation; motif discovery; muscle development; regulatory gene network; residual feed intake.
Figures


References
-
- Aerts S., Quan X. J., Claeys A., Sanchez M. N., Tate P., Yan J., et al. (2010). Robust target gene discovery through transcriptome perturbations and genome-wide enhancer predictions in drosophila uncovers a regulatory basis for sensory specification. PLoS Biol. 8:e1000435. 10.1371/journal.pbio.1000435.g001 - DOI - PMC - PubMed
-
- Archer J. A., Richardson E. C., Herd R. M., Arthur P. F. (1999). Potential for selection to improve efficiency of feed use in beef cattle: a review. Aust. J. Agric. Res. 50 147–161. 10.1071/A98075 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

