Population Pharmacogenomics for Precision Public Health in Colombia
- PMID: 30967898
- PMCID: PMC6439339
- DOI: 10.3389/fgene.2019.00241
Population Pharmacogenomics for Precision Public Health in Colombia
Abstract
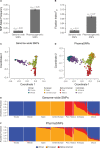
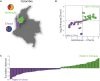
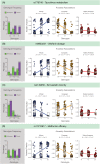
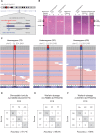
While genomic approaches to precision medicine hold great promise, they remain prohibitively expensive for developing countries. The precision public health paradigm, whereby healthcare decisions are made at the level of populations as opposed to individuals, provides one way for the genomics revolution to directly impact health outcomes in the developing world. Genomic approaches to precision public health require a deep understanding of local population genomics, which is still missing for many developing countries. We are investigating the population genomics of genetic variants that mediate drug response in an effort to inform healthcare decisions in Colombia. Our work focuses on two neighboring populations with distinct ancestry profiles: Antioquia and Chocó. Antioquia has primarily European genetic ancestry followed by Native American and African components, whereas Chocó shows mainly African ancestry with lower levels of Native American and European admixture. We performed a survey of the global distribution of pharmacogenomic variants followed by a more focused study of pharmacogenomic allele frequency differences between the two Colombian populations. Worldwide, we found pharmacogenomic variants to have both unusually high minor allele frequencies and high levels of population differentiation. A number of these pharmacogenomic variants also show anomalous effect allele frequencies within and between the two Colombian populations, and these differences were found to be associated with their distinct genetic ancestry profiles. For example, the C allele of the single nucleotide polymorphism (SNP) rs4149056 [Solute Carrier Organic Anion Transporter Family Member 1B1 (SLCO1B1)∗5], which is associated with an increased risk of toxicity to a commonly prescribed statin, is found at relatively high frequency in Antioquia and is associated with European ancestry. In addition to pharmacogenomic alleles related to increased toxicity risk, we also have evidence that alleles related to dosage and metabolism have large frequency differences between the two populations, which are associated with their specific ancestries. Using these findings, we have developed and validated an inexpensive allele-specific PCR assay to test for the presence of such population-enriched pharmacogenomic SNPs in Colombia. These results serve as an example of how population-centered approaches to pharmacogenomics can help to realize the promise of precision medicine in resource-limited settings.
Keywords: Antioquia; Chocó; Colombia; admixture; genetic ancestry; pharmacogenetics; pharmacogenomics; precision medicine.
Figures
References
-
- Andrews S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [accessed September 2017].
-
- Appelbaum N. P. (2016). Mapping the Country of Regions: The Chorographic Commission of Nineteenth-Century Colombia. Chapel Hill, NC: University of North Carollina Press.
-
- Bachtiar M., Lee C. G. (2013). Genetics of population differences in drug response. Curr. Genet. Med. Rep. 1 162–170.
LinkOut - more resources
Full Text Sources

