Robust Estimation of Recent Effective Population Size from Number of Independent Origins in Soft Sweeps
- PMID: 30968124
- PMCID: PMC6736332
- DOI: 10.1093/molbev/msz081
Robust Estimation of Recent Effective Population Size from Number of Independent Origins in Soft Sweeps
Abstract
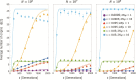
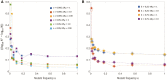
Estimating recent effective population size is of great importance in characterizing and predicting the evolution of natural populations. Methods based on nucleotide diversity may underestimate current day effective population sizes due to historical bottlenecks, whereas methods that reconstruct demographic history typically only detect long-term variations. However, soft selective sweeps, which leave a fingerprint of mutational history by recurrent mutations on independent haplotype backgrounds, holds promise of an estimate more representative of recent population history. Here, we present a simple and robust method of estimation based only on knowledge of the number of independent recurrent origins and the current frequency of the beneficial allele in a population sample, independent of the strength of selection and age of the mutation. Using a forward-time theoretical framework, we show the mean number of origins is a function of θ=2Nμ and current allele frequency, through a simple equation, and the distribution is approximately Poisson. This estimate is robust to whether mutants preexisted before selection arose and is equally accurate for diploid populations with incomplete dominance. For fast (e.g., seasonal) demographic changes compared with time scale for fixation of the mutant allele, and for moderate peak-to-trough ratios, we show our constant population size estimate can be used to bound the maximum and minimum population size. Applied to the Vgsc gene of Anopheles gambiae, we estimate an effective population size of roughly 6×107, and including seasonal demographic oscillations, a minimum effective population size >3×107, and a maximum <6×109, suggesting a mean ∼109.
Keywords: Anopheles; demographic oscillations; effective population size; recurrent mutation; soft sweeps.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures