Alterations in Striatal microRNA-mRNA Networks Contribute to Neuroinflammation in Multiple System Atrophy
- PMID: 30968343
- PMCID: PMC7216788
- DOI: 10.1007/s12035-019-1577-3
Alterations in Striatal microRNA-mRNA Networks Contribute to Neuroinflammation in Multiple System Atrophy
Abstract
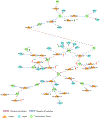
Multiple systems atrophy (MSA) is a rare neurodegenerative disorder characterized by the accumulation of α-synuclein in glial cells and neurodegeneration in the striatum, substantia nigra, and cerebellum. Aberrant miRNA regulation has been associated with neurodegeneration, including alterations of specific miRNAs in brain tissue, serum, and cerebrospinal fluid from MSA patients. Still, a causal link between deregulation of miRNA networks and pathological changes in the transcriptome remains elusive. We profiled ~ 800 miRNAs in the striatum of MSA patients in comparison to healthy individuals to identify specific miRNAs altered in MSA. In addition, we performed a parallel screening of 700 transcripts associated with neurodegeneration to determine the impact of miRNA deregulation on the transcriptome. We identified 60 miRNAs with abnormal levels in MSA brains that are involved in extracellular matrix receptor interactions, prion disease, inflammation, ubiquitin-mediated proteolysis, and addiction pathways. Using the correlation between miRNA expression and the abundance of their known targets, miR-124-3p, miR-19a-3p, miR-27b-3p, and miR-29c-3p were identified as key regulators altered in MSA, mainly contributing to neuroinflammation. Finally, our study also uncovered a potential link between Alzheimer's disease (AD) and MSA pathologies that involves miRNAs and deregulation of BACE1. Our results provide a comprehensive appraisal of miRNA alterations in MSA and their effect on the striatal transcriptome, supporting that aberrant miRNA expression is highly correlated with changes in gene transcription associated with MSA neuropathology, in particular those driving inflammation, disrupting myelination, and potentially impacting α-synuclein accumulation via deregulation of autophagy and prion mechanisms.
Keywords: Alpha-synuclein; Inflammation; Multiple systems atrophy; Neurodegeneration; Transcription; microRNA.
Figures





References
-
- Spillantini MG, Crowther RA, Jakes R, Cairns NJ, Lantos PL, Goedert M (1998) Filamentous alpha-synuclein inclusions link multiple system atrophy with Parkinson’s disease and dementia with Lewy bodies. Neurosci Lett 251(3):205–208 - PubMed
-
- Wakabayashi K, Hayashi S, Kakita A, Yamada M, Toyoshima Y, Yoshimoto M, Takahashi H (1998) Accumulation of alpha-synu- clein/NACP is a cytopathological feature common to Lewy body disease and multiple system atrophy. Acta Neuropathol 96(5):445–452 - PubMed
-
- Wakabayashi K, Yoshimoto M, Tsuji S, Takahashi H (1998) Alpha- synuclein immunoreactivity in glial cytoplasmic inclusions in multiple system atrophy. Neurosci Lett 249(2–3):180–182 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

