Hepatic Insulin Clearance: Mechanism and Physiology
- PMID: 30968756
- PMCID: PMC6734066
- DOI: 10.1152/physiol.00048.2018
Hepatic Insulin Clearance: Mechanism and Physiology
Abstract
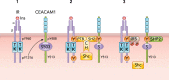
Upon its secretion from pancreatic β-cells, insulin reaches the liver through the portal circulation to exert its action and eventually undergo clearance in the hepatocytes. In addition to insulin secretion, hepatic insulin clearance regulates the homeostatic level of insulin that is required to reach peripheral insulin target tissues to elicit proper insulin action. Receptor-mediated insulin uptake followed by its degradation constitutes the basic mechanism of insulin clearance. Upon its phosphorylation by the insulin receptor tyrosine kinase, carcinoembryonic antigen-related cell adhesion molecule 1 (CEACAM1) takes part in the insulin-insulin receptor complex to increase the rate of its endocytosis and targeting to the degradation pathways. This review summarizes how this process is regulated and how it is associated with insulin-degrading enzyme in the liver. It also discusses the physiological implications of impaired hepatic insulin clearance: Whereas reduced insulin clearance cooperates with increased insulin secretion to compensate for insulin resistance, it can also cause hepatic insulin resistance. Because chronic hyperinsulinemia stimulates hepatic de novo lipogenesis, impaired insulin clearance also causes hepatic steatosis. Thus impaired insulin clearance can underlie the link between hepatic insulin resistance and hepatic steatosis. Delineating these regulatory pathways should lead to building more effective therapeutic strategies against metabolic syndrome.
Figures

References
-
- Al-Share QY, DeAngelis AM, Lester SG, Bowman TA, Ramakrishnan SK, Abdallah SL, Russo L, Patel PR, Kaw MK, Raphael CK, Kim AJ, Heinrich G, Lee AD, Kim JK, Kulkarni RN, Philbrick WM, Najjar SM. Forced hepatic overexpression of CEACAM1 curtails diet-induced insulin resistance. Diabetes 64: 2780–2790, 2015. doi: 10.2337/db14-1772. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

