Maturity2, a novel regulator of flowering time in Sorghum bicolor, increases expression of SbPRR37 and SbCO in long days delaying flowering
- PMID: 30969968
- PMCID: PMC6457528
- DOI: 10.1371/journal.pone.0212154
Maturity2, a novel regulator of flowering time in Sorghum bicolor, increases expression of SbPRR37 and SbCO in long days delaying flowering
Abstract
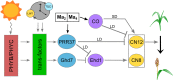
Sorghum bicolor is a drought-resilient facultative short-day C4 grass that is grown for grain, forage, and biomass. Adaptation of sorghum for grain production in temperate regions resulted in the selection of mutations in Maturity loci (Ma1 -Ma6) that reduced photoperiod sensitivity and resulted in earlier flowering in long days. Prior studies identified the genes associated with Ma1 (PRR37), Ma3 (PHYB), Ma5 (PHYC) and Ma6 (GHD7) and characterized their role in the flowering time regulatory pathway. The current study focused on understanding the function and identity of Ma2. Ma2 delayed flowering in long days by selectively enhancing the expression of SbPRR37 (Ma1) and SbCO, genes that co-repress the expression of SbCN12, a source of florigen. Genetic analysis identified epistatic interactions between Ma2 and Ma4 and located QTL corresponding to Ma2 on SBI02 and Ma4 on SBI10. Positional cloning and whole genome sequencing identified a candidate gene for Ma2, Sobic.002G302700, which encodes a SET and MYND (SYMD) domain lysine methyltransferase. Eight sorghum genotypes previously identified as recessive for Ma2 contained the mutated version of Sobic.002G302700 present in 80M (ma2) and one additional putative recessive ma2 allele was identified in diverse sorghum accessions.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Quinby JR. The Genetic Control of Flowering and Growth in Sorghum. Adv Agron. 1974;25:125–62.
-
- Olson SN, Ritter K, Rooney W, Kemanian A, McCarl BA, Zhang Y, et al. High biomass yield energy sorghum: developing a genetic model for C4 grass bioenergy crops. Biofuels, Bioprod Biorefining. 2012;6(3):246–56.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

