Epidemiological and genetic characterization of pH1N1 and H3N2 influenza viruses circulated in MENA region during 2009-2017
- PMID: 30971204
- PMCID: PMC6458790
- DOI: 10.1186/s12879-019-3930-6
Epidemiological and genetic characterization of pH1N1 and H3N2 influenza viruses circulated in MENA region during 2009-2017
Abstract
Background: Influenza surveillance is necessary for detection of emerging variants of epidemiologic and clinical significance. This study describes the epidemiology of influenza types A and B, and molecular characteristics of surface glycoproteins (hemagglutinin [HA] and neuraminidase [NA]) of influenza A subtypes: pH1N1 and H3N2 circulated in Arabian Gulf, Levant and North Africa regions during 2009-2017.
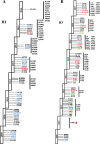
Methods: Analysis of phylogenetics and evolution of HA and NA genes was done using full HA and NA sequences (n = 1229) downloaded from Influenza Research Database (IRD).
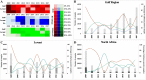
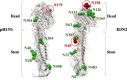
Results: In total, 130,354 influenza positive cases were reported to WHO during study period. Of these, 50.8% were pH1N1 positive, 15.9% were H3N2 positives and 17.2% were influenza B positive. With few exceptions, all three regions were showing the typical seasonal influenza peak similar to that reported in Northern hemisphere (December-March). However, influenza activity started earlier (October) in both Gulf and North Africa while commenced later during November in Levant countries. The molecular analysis of the HA genes (influenza A subtypes) revealed similar mutations to those reported worldwide. Generally, amino acid substitutions were most frequently found in head domain in H1N1 pandemic viruses, while localized mainly in the stem region in H3N2 viruses. Expectedly, seasons with high pH1N1 influenza activity was associated with a relatively higher number of substitutions in the head domain of the HA in pH1N1 subtype. Furthermore, nucleotide variations were lower at the antigenic sites of pH1N1 viruses compared to H3N2 viruses, which experienced higher variability at the antigenic sites, reflecting the increased immunological pressure because of longer circulation and continuous vaccine changes. Analysis of NA gene of pH1N1 viruses revealed sporadic detections of oseltamivir-resistance mutation, H275Y, in 4% of reported sequences, however, none of NAI resistance mutations were found in the NA of H3N2 viruses.
Conclusions: Molecular characterization of H1N1 and H3N2 viruses over 9 years revealed significant differences with regard to position and function of characterized substitutions. While pH1N1 virus substitutions were mainly found in HA head domain, H3N2 virus substitutions were mostly found in HA stem domain. Additionally, more fixed substitutions were encountered in H3N2 virus compared to larger number of non-fixed substitutions in pH1N1.
Keywords: H3N2; Hemagglutinin; Influenza epidemics; Molecular evolution; Neuraminidase; Phylogeny; pH1N1.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
Hadi Yassine is a member of the editorial board of the journal. Otherwise, the authors declare they have no conflict of interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


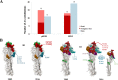
References
-
- World Health Organization- Influenza fact sheet. Influenza (Seasonal) Fact sheet. 2017 [cited 2018 February]; Available from: http://www.who.int/news-room/detail/14-12-2017-up-to-650-000-people-die-...
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

