Age-specific differences in the dynamics of protective immunity to influenza
- PMID: 30971703
- PMCID: PMC6458119
- DOI: 10.1038/s41467-019-09652-6
Age-specific differences in the dynamics of protective immunity to influenza
Abstract
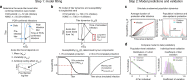
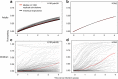
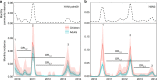
Influenza A viruses evolve rapidly to escape host immunity, causing reinfection. The form and duration of protection after each influenza virus infection are poorly understood. We quantify the dynamics of protective immunity by fitting individual-level mechanistic models to longitudinal serology from children and adults. We find that most protection in children but not adults correlates with antibody titers to the hemagglutinin surface protein. Protection against circulating strains wanes to half of peak levels 3.5-7 years after infection in both age groups, and wanes faster against influenza A(H3N2) than A(H1N1)pdm09. Protection against H3N2 lasts longer in adults than in children. Our results suggest that influenza antibody responses shift focus with age from the mutable hemagglutinin head to other epitopes, consistent with the theory of original antigenic sin, and might affect protection. Imprinting, or primary infection with a subtype, has modest to no effect on the risk of non-medically attended infections in adults.
Conflict of interest statement
B.J.C. has received research funding from Sanofi, and honoraria from Sanofi and Roche. The remaining authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

